Ube2W - figshare
advertisement

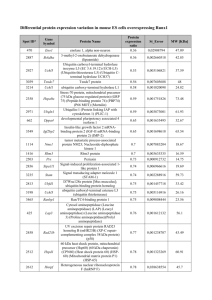
Deciphering the substrate specificity of ubiquitin conjugating enzymes Fábio M. Marques Madeira Supervisor: Professor Ronald T. Hay 24th July 2013 Protein ubiquitylation Hochstrasser, M. (2009) Nature 458, 422–9 1 The ubiquitin modification cascade RNF4 STUbL having a key role in DNA damage response Woelk, et al. (2007) Cell Division 2:11 2 RNF4 RING bond to ubiquitin-loaded UbcH5a Plechanovová, et al. (2012) Nature 489, 115–20 3 RNF4 RING bond to ubiquitin-loaded UbcH5a ε-amino group of lysine Tetrahedral transition state intermediary pKa 10.5 ± 1.1 Plechanovová, et al. (2012) Nature 489, 115–20 4 Ube2W conjugates ubiquitin to α-amino groups of protein N-termini α-amino group of the substrate N-terminus pKa 7.7 ± 0.5 Tatham, et al. (2013) The Biochemical Journal 453, 137–45 5 Aims Investigate what are the features of the active site of UbcH5a and Ube2W that enable them do discriminate between N-terminal α-amino groups and Lys εamino groups 1. Sequence and structure-informed mutational analysis of key residues 2. Protein expression and purification of the mutant proteins 3. Biochemical characterization of the proteins and in vitro ubiquitin conjugation assays 6 Structural analysis of UbcH5a and Ube2W Helix 1 D117 N * N77 Helix 2 UbcH5a (4AP4:E) Ube2W (2A7L:A) Ube2W model (I-TASSER) Ube2W model (Phyre2) Ubiquitin (4AP4:F) 7 Multiple alignment analysis of UbcH5a and Ube2W Helix 1 UbcH5a~Ubiquitin (4AP4:E~F) Helix 2 Model of Ube2W~Ubiquitin (4AP4:F) 8 Multiple alignment analysis of UbcH5a and Ube2W Helix 1 Helix 2 UbcH5a Ube2W M1 – N77H M2 – P115K/D116R/D117R M3 – M1/M2 M4 – D112S/insC/P113K/N114E/P115K/D116R/D117R M5 – M1/M4 M1 – H94N M2 – K133P/R134D/R135D M3 – M1/M2 M4 – S129D/delC130/K131P/E132N/K133P/R134D/R135D M5 – M1/M4 8 Protein expression and purification Site-directed mutagenesis 18 Mutant strand synthesis DpnI digestion ArcticExpress (DE3) Rosetta (DE3) E. coli BL21 (DE3) kDa wt M1 M2 M3 M4 M5 - + - + - + - + - + - + IPTG M2 M3 M4 M5 M2 M3 M4 M5 - + - + - + - + - + - + - + - + IPTG 75 - 75 - Cpn60 His6-UbcH5a Cpn10 50 - 50 - 37 - 37 25 20 - kDa DNA sequencing His6-UbcH5a 25 20 - 15 - 15 - 10 - 10 - 9 Protein expression and purification Site-directed mutagenesis 18 Mutant strand synthesis DpnI digestion E. coli Rosetta (DE3) E. coli BL21 (DE3) kDa wt M1 M2 M3 M4 M5 - + - + - + - + - + - + IPTG M2 M3 M4 M5 - + - + - + - + - + - + IPTG 37 - 37 - 10 - M1 50 - 50 - 15 - kDa wt 75 - 75 - 25 20 - DNA sequencing His6-UbcH5a 25 20 - His6-Ube2W 15 10 - 9 Protein expression and purification Lyse cells Wash Elute Expression vector Ni-NTA resin UbcH5a wt and M1-3 kDa S F B W His6-tagged proteins kDa 75 50 37 - 25 - C S F B W E T His6-UbcH5a UbcH5a 15 - kDa 75 - 75 - 50 - 50 - 37 - 37 - 25 20 - His6-Ube2W Ube2W 15 - 10 C – Cell suspension S – Supernatant His6-tag F – Flow-through B – First wash C S F B W His6-tag W – Second wash E – Elution E T 25 20 15 - 10 - Purified proteins Ube2W M2-5 Ube2W wt and M1 E T 20 - Cleavage with TEV protease 10 - His6-Ube2W Ube2W His6-tag T – After His6-tag cleavage with TEV 10 Protein expression and purification Gel filtration on a HiLoad 16/60 Superdex 75 pg Ube2W M2-5 C S F B W E T kDa 250 75 50 37 - 25 20 15 10 - His6-Ube2W Ube2W Absorvance at 280 nm (mAU) kDa 1 2 3 4 5 6 7 8 9 10 11 12 13 20 15 - 200 150 100 50 7 6 1 13 His6-tag 0 50 55 60 65 70 75 80 85 Elution volume (ml) Equilibrium of moners and dimers Vittal, et al. (2013) Cell Biochemistry and Biophysics 13, 9633-5 11 The ability of mutant proteins to form E2~Ub thioester bonds Reaction mix: E1 + E2 + Ub + ATP Ube2W UbcH5a wt kDa 25 - 20 - Non-reducing SDS-PAGE M1 M2 wt M3 Time kDa 25 - UbcH5a~Ub UbcH5a 20 - 15 - Ubiquitin M3 M4 M5 Time Ube2W~Ub Ube2W Ubiquitin Ube2W Ubiquitin 25 - 25 - 20 - 20 UbcH5a 15 - 15 10 - M2 10 - 10 - Reducing SDS-PAGE 15 - M1 Ubiquitin 10 - M1 – N77H M2 – P115K/D116R/D117R M3 – M1/M2 12 pH titration analysis of UbcH5a and Ube2W Reaction mix: + E1 Peptide-His6-SUMO-2x4 6.5 7.0 7.5 8.0 E3 + Ub + ATP + E2 8.5 9.0 9.5 N SUMO-2 SUMO-2 SUMO-2 SUMO-2 His6 10.0 10.5 11.0 pH Time kDa UbcH5a UbcH5a N77H 75 50 - His6-SUMO-2x4~Ub His6-SUMO-2x4 75 - 50 - 6.5 Ube2W 7.0 7.5 8.0 8.5 9.0 9.5 10.0 10.5 11.0 His6-SUMO-2x4~Ub His6-SUMO-2x4 pH kDa Time 75 - 50 - 75 - Ube2W M3 + 50 - His6-SUMO-2x4~Ub His6-SUMO-2x4 His6-SUMO-2x4~Ub His6-SUMO-2x4 M3 – H94N/ K133P/R134D/R135D 13 Conclusions 1. Key residues in the active site of Ube2W are different from most of the conserved E2s 2. Ube2W shows an equilibrium of monomers and dimers that does not rely on the C-terminus 3. Most of the mutant proteins can still form a thioester bond with ubiquitin, although their ability to modify a poly-SUMO2 substrate is affected 4. Ube2W shows pH-dependent activity at pH below 9.0 14 Future work 1. Try to overcome low expression of UbcH5a mutants by DNA synthesis of the constructs with codon optimization 2. Investigate what are the key features of N-terminal amino groups modified by Ube2W, using N-terminal modified substrates (myc-tag, in vitro carbamylation, etc.) 3. Try solving the structure of RNF4-Ube2W~Ubiquitin mutating the catalytic Cys to Lys to form an isopeptide linkage 15 Acknowledgements Professor Ronald T. Hay Anna Plechanovová Ellis Jaffray Linnan Shen Mike Tatham … all members of the Hay group!