Cytological Studies Of Maize [Zea Mays L.] and Teosinte
advertisement
![Cytological Studies Of Maize [Zea Mays L.] and Teosinte](http://s3.studylib.net/store/data/009550557_1-3a9d1be429a9d302239ace39bdea9ce1-768x994.png)
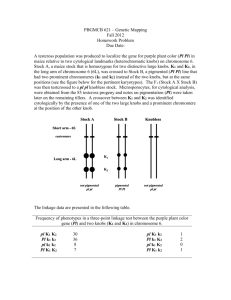
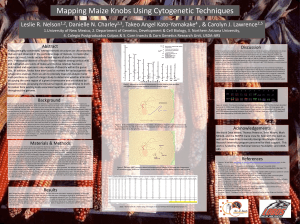
Positioning Maize Knobs Relative to B73 Maize Sequence Danielle N. Charley1&2, Leslie R. Nelson2&3 , Ethalinda Cannon2, Takeo Angel Kato Y4, and Carolyn J. Lawrence2&5 1Northern Arizona University, Flagstaff, AZ USA; 2Iowa State University, Ames, IA USA; 3University of New Mexico, Albuquerque, NM USA; 4Colegio Abstract: de Postgraduados, Chapingo, MEXICO; 5USDA-ARS CICGRU, Ames, IA USA Graph 4 Table 1 In the past knobs have been used to characterize the diversity of maize accessions and have been key in understanding genetic crossing over in maize as well as for other problems. Over time, they have become less widely utilized. During the assembly of the maize genome sequence, some repeat rich regions were removed which included some of the knob sequences. Because most knob sequence has been removed an objective of this research is to find where the knobs might be located relative to sequence. Regardless of where sequence has been removed, there remain some copies of the repeats. Using the MaizeGDB website and the tools offered within the site, I found regions where knob repeats are high, indicating that the knob itself may reside in that region. This research will aid researchers in using the maize genome assembly by alerting them to regions of the assemblies where sections of the genome sequence may be missing. Background: Maize has 10 chromosomes, on some of these chromosomes are heterochromatic regions called knobs. A heterochromatic region is a place in the genome where the DNA is tightly compacted and mainly consists of repeats. These regions contain the 180-bp repeat and the 350-bp TR-1 repeat (Peacock et al., 1981 and Ananiev et al., 1998). Studies have shown that there may be some association between knob constitution and phenotypic characteristics including yield (Wellhausen and Prywer 1954; Moll et al., 1972; and Chughtai and Steffensen 1987) . Knobs also were used by Barbara McClintock as cytological markers in her seminal discovery that links genetic crossing over with physical crossovers observed in chromosomes (Creighton, H., and McClintock 1931). There has been little research done which associates knob locations with the genomic sequence. TA Kato Y has been studying maize knobs for many years. Figure 1 is an idiogram of the maize karyotype, showing where he has observed knobs to exist (Kato unpublished). The goal of the project was to mark positions on the genome where the knobs may reside. Data TA Kato Y collected for chromosome 1 from various maize lines as well as the conversion of micrometer measurements to cytological map units for those data. Figure 2 Graph 1 Conclusions: Researchers understand the maize genome in many ways: cytologically, genetically, and sequentially. Placing knob data onto the various maps on all map types will help researchers to understand their placement across all major paradigms of biological understanding. Because all the work done here is inferential, the knob locations relative to the genome sequence represent hypotheses for location and must be further tested. This work serves as a framework for understanding where knobs may lie relative to markers mapped to the maize genome sequence. Materials and Methods: There are three different maps types used in this project: a genetic map, a sequence map, and a cytological map. All three of these have their own units: centiMorgan (cM), base pair (bp), and centiMcClintock (cMC). Each map type is collinear but each unit type is different. A cM is a statistical measure of distance based on segregation of traits in offspring of two inbred parents. A bp is a single nucleotide in a DNA sequence. A cMC is the percentage distance of a locus from the centromere to the end of the locus arm. Cytological representation of the knobs in the first sample on chromosomes 1-10 given TA Kato Y’s data. B73 RefGen_v1- reference genome assembly for this research, version 1 assembly of the B73 maize genome (Schnable et al., 2009). This graph is based on the IBM2 2008 Neighbors Map from MaizeGDB (cM). This is a genetic map showing where knobs might be using genetic data rather than cytological measurements. The IBM2008 map is a very common genetic map used in maize genetics This representation was created by drawing the genetic length of the chromosome, finding the repeat in MaizeGDB and reporting the genetic coordinates, then centromeres were added. Although only knob locations are shown here, many other loci including genes are available on the full IBM2 2008 Neighbors map. MaizeGDB- Maize Genetics and Genomics Database, used for data and visual analysis (Sen et al., 2009). BLAST- Used to align knob repeat sequence on B73 genome assembly (Altschul et al., 1997). Graph 2 Graph 3 CViT- Used for creating map images (Cannon and Cannon, 2005). GBrowse- A genome browser used for viewing features on genomic sequence (Stein et al., 2002). Locus Lookup- Searches for genomic coordinates of a locus (Andorf et al., 2010). IBM2 2008 Neighbors map- Provided genetic coordinates for knobs (Schaeffer et al., 2008). 180 bp and TR-1- repeat sequence used to identify knob regions (GenBank). Results: Figure 1 Sequence map (bp) created using MaizeGDB and Locus Pair Lookup. Locus Pair Lookup finds knobs on the different chromosomes and determines ranges for their locations based upon probe locations on the genome assembly and/or loci that are on either side of the knob on genetic maps. Those coordinates are used to approximate where the knob might be. The red are the ranges of where a knob might be and the black are the centromeres. Karyotype from T.A. Kato Y showing knob positions on chromosomes 1-10. Sequence map (bp) with full BLAST results. Repeats within 500 bp of each other are collapsed into regions. This map was created using the BLAST software. We took the sequence for the 180 base pair repeat and the TR-1 repeat and used the command line version of BLAST to find matches across the entire maize genome assembly. From this we massaged the data based upon percent identity and only show matches that were 90% identical or higher. We then converted those results into a gff file. The red regions are the BLAST hits and the black are the centromeres. Sequence map (bp) generated using GBrowse and BLAST to estimate where knobs are located. We used BLAST to search for clusters of knob repeats and uploaded hits to MaizeGDB’s instance of Gbrowse, which is a common software package for genome visualization. In this representation there could be errors because during the sequencing process and again for assembly, the Maize Genome Sequencing Consortium intentionally removed repetitive elements. This was done for sequencing because the goal was to sequence only genic regions of the genome. For assembly, repetitive elements were removed because it is difficult to correctly assemble highly repetitive regions correctly. The red regions are the knob hits that were found and the black regions represent centromeres. References: 1. Chughtai, S.R. and Steffensen, D.M. (1987) Maize Genetics Cooperation News Letter 61, 98-99. 2. Creighton, H., and McClintock, B. 1931 A correlation of cytological and genetical crossing-over in Zea mays. PNAS 17:492–497. 3. Durbin, R., Eddy, S., Krough, A., and Mitchison, G. (1998). Biological Sequence Analysis: Probabilistic Models of Protein and Nucleic Acid. Cambridge University Press, New York,NY. 4. Kato, Takeo Angel. Cytological Studies Of Maize [Zea Mays L.] and Teosinte [Zea Mexican (Scrader) Kuntze] In Relation to Their Origin And Evolution. Massachusetts: University of Massachusetts, 1975. 5. Lawrence, Carolyn J., Seigfried, Trent E., Bass, Hank W., and Anderson, Lorinda K.(2006). Predicting Chromosomal Locations of Genetically Mapped Loci in Maize Using the Morgan2McClintock Translator. Genetics Society of America. 6. Moll, R.G., Hansen, W.D., Levings, C.S., and Ohta, Y. (1972) Crop Sci. 12, 585-589. 7. Peacock, W.J. Dennis, E.S., Rhoades, M.M., and Pryor, A.J. Highly Repeated DNA Sequence Limited to Knob Heterochromatin in Maize. PNAS USA 1981 pg. 4490. 8. Sen, TZ, Andorf, CM, Schaeffer, ML, Harper, LC, Sparks, ME, Duvick, J, Brendel, VP, Cannon, E, Campbell, DA, Lawrence, CJ. (2009) MaizeGDB becomes 'sequence-centric' Database. 2009:Vol. 2009:bap020. 9. Wellhausen, E.J. and Prywer, C. (1954) Agron J. 46,507-511. 10.Sen, TZ, Andorf, CM, Schaeffer, ML, Harper, LC, Sparks, ME, Duvick, J, Brendel, VP, Cannon, E, Campbell, DA, Lawrence, CJ. (2009) MaizeGDB becomes 'sequence-centric' Database. 2009:Vol. 2009:bap020. 11.Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ Nucleic Acids Res. 1997 Sep 1;25(17):3389-402. 12.Cannon, EK; Cannon SB. (2005) CViT: Chromosome Visualization Tool, Unpublished 13.The generic genome browser: a building block for a model organism system database. Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, Lewis S. Genome Res. 2002 Oct;12(10):1599-610. 14.The Locus Lookup tool at MaizeGDB: identification of genomic regions in maize by integrating sequence information with physical and genetic maps. Andorf CM, Lawrence CJ, Harper LC, Schaeffer ML, Campbell DA, Sen TZ. Bioinformatics. 2010 Feb 1;26(3):434-6. 15.Schaeffer (Polacco), ML; Sanchez-Villeda, H; Coe, E. (2008) IBM2 2008 Neighbors, Unpublished 16.GenBank accessions: DQ186871.1, M32528.1 Acknowledgments: As a Native American Outreach Program participant, I am very grateful to the sponsors that have made this program possible. A special thanks to my Mentors and Graduate Student Mentors who have put the time and effort into helping me grow as researcher and student. List of Sponsors: Carolyn Lawrence, Ethalinda Cannon, Trent Moore, Mary De Baca, Aurelio Curbelo, Jovaughn Barnard, Dustin Thunder Hawk, Ranelle White Buffalo, George Washington Carver Internship, National Science Foundation, USDAARS, and Iowa State University.