Chapter 5 Membranes
advertisement

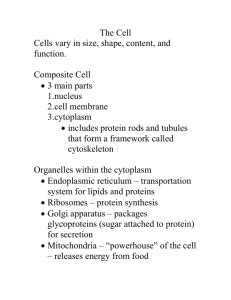
Chapter 5 Membranes I. The Structure of Membranes (5.1) A. The fluid mosaic model shows proteins embedded in a fluid lipid bilayer 1. Phospholipid a. Hydrophilic head (glycerol and phosphate group make it polar) b. Hydrophobic tail (2 fatty acid chains are nonpolar) 2. Globular proteins a. proteins w/ nonpolar segments and polar ends to fit in phospholipid bilayer. 3. Fluid mosaic model a. random arrangement of proteins “floating” in or on the fluid lipid bilayer. http://www.susanahalpine.com/anim/Life/memb.htm B. Cellular membranes consist of four component groups 1. Phospholipid bilayer: 2 layers of phospholipids with hydrophobic tails pointed inward. 2. Transmembrane proteins (aka intergral membrane proteins): allow substances to pass through membrane. 3. Interior protein network: proteins in the membrane that help support cell shape. a. Peripheral membrane proteins: proteins that are not part of the membrane structure but help control cell movement. 4. Cell surface markers: a. Glycoproteins and glycolipids act as cell identity markers (IDs) • Lipid rafts: sections of the cell membrane that function as signal receptors and involved in cell movement tightly packed by cholesterol. C. Electron microscopy has provided structural evidence Transmission electron microscopes (TEM) and scanning electron microscopes (SEM) used to study the plasma membrane. II. Phospholipids: The Membrane’s Foundation (5.2) A. Phospholipids spontaneously form bilayers 1. Hydrophilic phosphate group facing out 2. Hydrophobic nonpolar fatty acids face in 3. Closely packed hydrophobic tails prevent water soluble substances from passing through. B. The phospholipid bilayer is fluid 1. Hydrogen bonds help hold membrane together. 2. Weak forces b/t phospholipids allow movement of proteins and lipids. C. Membrane fluidity can change 1. Saturated fats and cooler temps make the membrane less fluid (solid) b/c lipids are tightly packed. 2. Unsaturated fats and higher temps make the membrane more fluid b/c lipids are more loosely packed. • http://telstar.ote.cmu.edu/Hughes/tutorial/c ellmembranes/ III. Proteins: Multifunctional Components (5.3) A. Proteins and protein complexes perform key functions 1. Transporters: move substances across membrane in carriers or channels 2. Enzymes: carry out chemical reactions 3. Cell surface receptors: detect messages 4. Cell surface identity markers: ID 5. Cell-to-cell adhesion proteins: glue cells together 6. Attachments to the cytoskeleton B. Structural features of membrane proteins 1. The anchoring of proteins in the bilayer: nonpolar sections of protein tied to membrane. α-helix or -sheets 2. Transmembrane domains: area or areas of protein anchored to nonpolar lipid bilayer. α-helix 3. Pores: proteins that form pipelike holes to allow passage for molecules -sheets IV. Passive Transport Across Membranes A. Passive transport: movement of substances in and out of the cell w/OUT using energy B. Concentration gradient: difference in concentration of a substance b/t the inside and outside of a cell. C. Transport can occur by simple diffusion 1. Diffusion: random movement of molecules from an area of high concentration to an area of lower concentration. • http://www.stolaf.edu/people/giannini/flash animat/transport/diffusion.swf • http://www.wiley.com/college/pratt/047139 3878/student/animations/membrane_trans port/index.html • NOTE: Nonpolar molecules like O2 and steroids have a much easier time moving across the lipid bilayer membrane. Few small polar molecules can cross very slowly • http://www.teachersdomain.org/resources/t dc02/sci/life/cell/membraneweb/assets/tdc 02_int_membraneweb/tdc02_int_membra neweb_swf.html D. Proteins allow membrane diffusion to be selective 1. Channel proteins: provide a passage for polar molecules 2. Carrier proteins: bind to molecule and help them cross membrane. 3. Selectively permeable: choose which molecules can cross. 4. Diffusion of ions through channels a. Ion channels: transport channel proteins move ions (i.e. K+, Ca+2, Cl-) across according to their concentration. b. Gated channels: channels proteins that open or close due to a stimulus. c. Membrane potential: difference in voltage across membrane causes channels to open or close. E. Carrier proteins and facilitated diffusion a. Facilitated diffusion: carrier proteins help move molecules across membrane according to the concentration difference. b. Saturation: at high concentrations, all carrier proteins will move molecules across. http://highered.mcgrawhill.com/sites/0072495855/student_view0/ chapter2/animation__how_facilitated_diffu sion_works.html c. Facilitated diffusion is: - specific to certain molecules - passive: high to low concentration - saturates: only moves as fast as # of carrier proteins available. 6. Facilitated diffusion in red blood cells C. Osmosis is the movement of water across membranes 1. Solvent: medium, usually water, in which solute is dissolved. 2. Solute: substance dissolved in solvent ***Solutes and solvents move by diffusion (high concentration to low concentration) 3. Osmosis: movement of water to area of higher solute concentration. a. Osmotic pressure: the concentration of all solutes in a solution. http://highered.mcgrawhill.com/sites/0072495855/student_view0 /chapter2/animation__how_osmosis_wor ks.html b. Hypertonic: higher solute concentration c. Hypotonic: lower solute concentration d. Isotonic: when two solutions have the same osmotic (solute) concentration. 4. Aquaporins: water channels • Movement of water is assisted by special water channels. • People with Nephrogenic Diabetes Insipidus have nonfunctional aquaporins and have trouble retaining water. 5. Osmotic pressure a. Hydrostatic pressure: pressure of cytoplasm pushing on cell membrane due to osmosis. a. Osmotic pressure: force to stop flow of water, depends on solute concentration inside and outside of cell. 6. Maintaining osmotic balance a. Extrusion: contractile vacuole pumps out water that constantly enters due to osmosis. b. Isosmotic Regulation: Increasing the internal solute concentration to match the environment. • http://trc.ucda vis.edu/biosci 10v/bis10v/we ek7/parameci um.mov c. Turgor/ Turgor pressure: plant cells are usually hypertonic to their environment to increase hydrostatic pressure. This increases the pressure of the cell membrane against the cell wall maintaining cell shape. • http://www.teachersdomain.org/resources/t dc02/sci/life/cell/membraneweb/assets/tdc 02_int_membraneweb/tdc02_int_membra neweb_swf.html V. Active Transport Across Membranes (5.5) A. Active transport : movement of substances UP CONCENTRATION GRADIENT that requires ENERGY from ATP. B. Active transport uses energy to move materials against a concentration gradient 1. Uniporters: carrier proteins that move only 1 type of molecule. 2. Symporters: carrier proteins that move 2 molecules in the same direction. 3. Antiporters: carrier proteins that move 2 molecules in opposite directions. 3 types of carrier proteins using active transport C. The sodium-potassium pump runs directly on ATP 1. Sodium-potassium pump: carrier protein that moves 3 sodium ions (Na+) out of the cell for every 2 potassium ions (K+) it moves in. It uses ATP to move these ions from areas of low concentration to areas of high concentration. http://highered.mcgrawhill.com/sites/0072495855/student_view0/chapter2/animation__how_the_ sodium_potassium_pump_works.html STEPS of Na+/K+ Pump 1. 3 Na+ bind to protein changing its conformation (shape). 2. Protein separates ATP to ADP and binds to phosphate group to becoming phosphorylated 3. Phosphorylated protein moves 3 Na+ to outside fluid 4. Protein has new conformation and binds to K+ from outside 5. Binding to K+ causes a conformation in protein to release phosphate group 6. The protein is now free of phosphate group and changes back to original conformation with a high affinity for 3 Na+ http://www.brookscole.com/chemistry_d/templates/student_resources/s hared_resources/animations/ion_pump/ionpump.html D. Coupled transport uses ATP indirectly 1. Coupled transport: the energy released to move a molecule down its concentration gradient is used to move a different molecule against its concentration gradient. EXAMPLE: • Sodium-potassium pump uses ATP to move sodium ions outside of cell where there is a higher concentration. • A symporter moves sodium ions into the cell and simultaneously moves glucose into the cell where there is a higher concentration. 2. Countertransport An antiporter moves one molecule, such as Na+, is moved down its concentration gradient. The energy is used to move another molecule, such as H+, in the opposite direction against its concentration gradient. Coupled transport and Countertransport • http://highered.mcgrawhill.com/olc/dl/120068/bio04.swf VI. Bulk Transport by Endocytosis and Exocytosis A. Bulk transport: process moving LARGE, POLAR molecules across membrane B. Bulk material enters the cell in vesicles 1. Endocytosis: plasma membrane takes in food particles and liquids. 2. Phagocytosis (eat): cell takes in an organism or large organic molecules (WBC take in bacteria) 3. Pinocytosis (drink): cell takes in liquid (mammalian egg cell take in nutrients) 3. Receptor-mediated endocytosis: molecules bind to receptors on the membrane. Clathrin proteins help to trap these molecules and form a vesicle. Molecules like LDLs enter through this process. C. Material can leave the cell by exocytosis 1. Exocytosis: movement of large materials out of the cell via vesicles. (moves cell wall materials, excess water, enzymes, hormones, neurotransmitters and waste). • http://highered.mcgrawhill.com/olc/dl/120068/bio02.swf