dv2
advertisement

Macromolecular Crystallography
and
Structural Genomics
– Recent Trends
Prof. D. Velmurugan
Department of Crystallography and Biophysics
University of Madras
Guindy Campus, Chennai – 25.
• Structural Genomics aims in identifying as many
new folds as possible.
• This eventually requires faster ways of
determining the three dimensional structures as
there are many sequences before us for which
structural information is not yet available.
• Although Molecular Replacement technique is
still used in Crystallography for solving
homologous structures, this method fails if there
is not sufficient percentage of homology.
• The Multiwavelength Anomalous Diffraction
(MAD) techniques have taken over the
conventional Multiple Isomorphous Replacement
(MIR) technique.
• With the advent of high energy synchrotron sources and
powerful detectors for the diffracted intensities,
developments in methodologies of macromolecular
structure determination, there is a steep increase in the
number of macromolecular structures determined and
on an average eight new structures are deposited in the
PDB every day and the total entries in the PDB is now
around 29,000.
• Instead of using the three wavelength strategies in MAD
experiments, the use of single wavelength anomalous
diffraction using Sulphur anomalous scattering is
recently proposed. This will reduce the data collection
time to 1/3rd.
• Also, the judicious use of the radiation damage during
redundant data measurements in second generation
synchrotron source and also during regular data
collection in the third generation synchrotron source has
been pointed out recently (RIP & RIPAS).
Protein Structure Determination
•
•
•
•
•
X-ray crystallography
NMR spectroscopy
Neutron diffraction
Electron microscopy
Atomic force microscopy
As the number of available amino acid
sequences exceeds far in number than the
number of available three-dimensional
structures, high-throughput is essential in
every aspect of X-ray crystallography.
Procedure
Protein Crystal
The 14 Bravais lattices
1: Triclinic
2: Monoclinic
(Blue numbers correspond o the crystal system)
The 14 Bravais lattices
3: Orthorhombic
(Blue numbers correspond to the crystal system)
The 14 Bravais lattices
4: Rhombohedral
5: Tetragonal
(Blue numbers correspond to the crystal system)
6: Hexagonal
The 14 Bravais lattices
7: Cubic
(Blue numbers correspond to the crystal system)
Synchrotron radiation
More intense X-rays at shorter wavelengths mean higher
resolution & much quicker data collection
Diffraction Apparatus
Diffraction Principles
nl = 2dsinq
The diffraction experiment
The amplitudes of the waves scattered by an atom to that of an single
electron
– atomic scattering factor
The amplitude of the waves scattered by all the atoms in a unit cell to
that of a single electron (The vector (amplitude and phase) representing the overall
scattering from a particular set of Bragg planes) | Fhkl | – structure factor
The structure factor magnitude F(hk/) is represented by the length
of a vector in the complex plane.
The phase angle a(hk/) is given by the angle. measured
counterclockwise, between the positive real axis and the vector F.
unit cell
F (h,k,l) = Vx=0 y=0 z=0 (x,y,z).exp[2I(hx + ky + lz)].dxdydz
A reflection
electron density
V = the volume of the unit cell
|Fhkl| = the structure-factor amplitude (proportional to the
square-root of reflection intensities)
ahkl = the phase associated with the structure-factor amplitude
|Fhkl|We can measure the amplitudes, but the phases are lost in
the experiment.
This is the phase problem.
Fourier Transform requires both
structure factors and phases
Electron density calculation
ρ
ΣΣΣ
π
α
Unknown
Patterson function
• Patterson space has the same dimension as
the real-space unit cell
• The peaks in the Patterson map are
expressed in fraction coordinates
• To avoid confusion, the x, z and z
dimensions of Patterson vector-space are
called (u, v, w).
What does Patterson function
represent?
• It represents a density map of the vectors
between scattering atoms in the cell
• Patterson density is proportional to the
squared term of scattering atoms, therefore,
the electron rich, i.e., heavy atoms,
contribute more to the patterson map than
the light atoms.
Patterson function –
no phase info required
Consider phaseless term (h, k, l, F2)
ΣΣΣ
P
No phase term
Patterson map
r =
hkl F (S) exp (-2i{r.S})
Direct space
Pu = hkl I (S) exp (-2i{u.S})
Density
and
position
P(u) =
Amplitudes
and
phases
cell rr+u d3r
I(S)=F*(S).F(S)=|F(S)|2
F (S) =
cell r exp (2i{r.S}) d3r
Fourier
transformation
Fourier
transformation
Patterson
map
Intensities
Reciprocal space
r =
hkl F (S) exp (-2i{r.S})
Patterson map with symmetry
Patterson map symmetry
Harker vectors
u, v, w
2x, 1/2, 2z
P(u) =
cell rr+u d3r
P21
x, y, z
-x, y+1/2, -z
Pu = hkl I (S) exp (-2i{u.S})
Diffracting a Cat
Diffraction
data with
phase
information
Real
Diffraction
Data
Reconstructing a Cat
FT
Easy
FT
Hard
The importance of phases
Phasing Methods
all assume some prior knowledge of the electron
density or structure
The Phase Problem
• Diffraction data only records intensity,
not phase information (half the
information is missing)
• To reconstruct the image properly you
need to have the phases (even approx.)
–
–
–
–
Guess the phases (molecular replacement)
Search phase space (direct methods)
Bootstrap phases (isomorphous replacement)
Uses differing wavelengths (anomolous disp.)
Acronyms for phasing techniques
•
•
•
•
•
•
•
MR
SIR
MIR
SIRAS
MIRAS
MAD
SAD
Direct methods
• Based on the positivity and atomicity of electron density that leads
to phase relationships between the (normalized) structure factors
(E).
• Used to solve small molecules structures
• Proteins upto ~1000 atoms, resolution better than 1.2 Å
• Used in computer programs (SnB, SHELXD SHARP) to find
heavy-atom substructure.
Jerome Karle and Herbert A. Hauptman
Nobel prize 1985 (chemistry)
Density modification procedures (e.g. solvent flattening and
averaging) can be carried out as part of a cyclic process
Dm cycle
P
h
a
s
e
sa
n
d
a
m
p
l
it
u
d
e
s
F
,
P
P
Fourier
transformation
Phase
combination
N
e
w
p
h
a
s
e
s
a
n
d F
,
P
c
a
l
c
P
c
a
l
c
a
m
p
l
i
t
u
d
e
s
l
e
c
t
r
o
n
(
r
)E
d
e
n
s
i
t
y
Map
modification
Inverse Fourier
transformation
(
r
)
m
o
d
M
o
d
i
f
i
e
d
e
l
e
c
t
r
o
n
d
e
n
s
i
t
y
m
a
p
Molecular Replacement (MR)
Used when there is a homology model available (sequence identity > 25%).
1. Orientation of the model in the new unit cell (rotation function)
2. Translation
Molecular Replacement (MR)
• MR works because the Fourier
transform works in both directions.
– Reflections
New Protein
Coordinates in PDB
model (density)
• Have to be careful of model bias
MR solution
Isomorphous replacement
• Why isomorphous replacement, making
heavy atom derivatives?
– Phase determination
• Calculating FH
FH= FPH-FP
If HA position is known, FH can be calculated from ρ(xH, yH,
zH) by inverse FT
• HA position determination – Patterson
function
HA shifts FP by FH
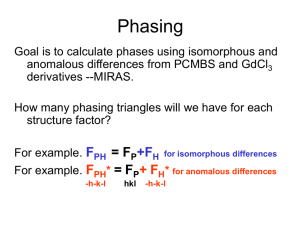
Isomorphous Replacement (SIR, MIR)
– Collect data on native crystals (no metals)
– Soak in heavy metal compounds into crystals,
go to specific sites in the unit cell.
• e.g. Hg, Pt, Au compounds
– The unit cell must remain isomorphous
– Collect data on the derivatives
– As a result, only the intensity of the
reflections changes but not the indices
– Measure the reflection intensity differences
between native and derivative data sets.
– Find the position of the heavy atoms in the
unit cell from the intensity differences.
• generate vector maps (Patterson maps)
Native and heavy-atom derivative
• |FP + HA| – |FP| = |FHA|
• Must have at least two heavy atom derivatives
• The main limitations in obtaining accurate
phasing from MIR is non isomorphism and
incomplete incorporation (low occupancy) of
the heavy atom compound.
diffraction patterns superimposed
and shifted vertically.
Note: intensity differences for
certain reflections.
Note: the identical unit cell
(reflection positions).
This suggests isomorphism.
Isomorphic HA derivatives only changes
the intensity of the diffraction but not the
indices of the reflections
Native crystal
HA derivative crystal
Once we have an heavy atom structure H(r), we can use this to calculate
FH(S). In turn, this allows us to calculate phases for FP and FPH for each
reflection.
Harker diagram
Harker construction for SIR
FP
PH(P)
FPH
-F H
FP
P
FPH
The phase probability
distribution shows that SIR
results in a phase
ambiguity
We can use a second derivative to resolve the phase ambiguity
Harker construction for
multiple isomorphous replacement (MIR)
MIR
PH(P)
FPH2
FP
P
-FH2
FPH
-F H
PH2(P)
P
P(P) = PH(P).PH2(P)
P
AS
Anomalous scattering leads to a breakdown of Friedel‘s law
FPH(S)
Anomalous
derivative
FPH(0)
FPH(-S)
Anomalous scattering data can also be used to solve the phase ambiguity
P+(P)
P
+
FP
-F +H'
+
PH
F
F*PH
P(P)
-F+H'' -F*H''
P
P(P) = P+(P).P(P)
P
Note that the anomalous differences are very
small; thus very accurate data are necessary
Of course, there are errors in the data, determination of heavy atom
positions etc.
Blow and Crick developed a model in which all errors are
associated with |FPH|obs
FH
FPH
FP
P
PH
The triangle formed by FP, FPH and FH
fails to close
The 'lack of closure error' is a function
of the calculated phase angle P
=
||
F
||
F
P
H
o
b
s
P
H
c
a
l
c
The phase probability P(P) is given by
(P)
P(P) = exp
2E2
2
The resulting phases have a
minimum error when the
best phase best, i.e. the centroid of
the phase distribution
best =
2
0 P( )d
P
P
P
is used instead of the
most probable phase.
The quality of the phases is
indicated by the figure of merit m
m=
most probable phase
0
2
P(P)exp(iP)dP
2
0 P(P)dP
m=1: 0o phase error
o
m=0.5: ~60 phase error
m=0: all phases equally probable
•
Steps
in
MAD
Introduce anomalous scatterer
– Incorporate SeMet in replace of Met
– Incorporate HA eg Hg, Pt, etc…
•
Take your crystals to a synchrotron beam-line
(tunable wavelength).
•
Collect data sets at 3 separate wavelengths:
the Se (or other HA) absorption peak, edge and
distant to the peak.
•
Measure the differences in Friedel mates to get
an estimate of the phases for the Se atoms.
– These differences are quite small so one need to
collect a lot of data (completeness, redundancy)
to get a good estimate of the error associated
with each measurement.
•
Use the Se positions to obtain phase estimates
for the protein atoms.
Atomic scattering factor: 3 terms
Advantages of MAD
• All data is collected from one crystal
– Perfect isomorphism
• Fast
• Easily interpretable electron density maps
obtained right away.
SAD
Single-wavelength anomalous diffraction (SAD)
phasing has become increasingly popular in protein
crystallography.
Two main steps –
1) obtaining the initial phases
2) improving the electron density map
calculated with initial phases.
• The essential point is to break the intrinsic phase ambiguity.
• Two kinds of phase information enables the discrimination of phase
doublets from SAD data prior to density modification.
From heavy atoms (expressed by Sim distribution)
From direct methods phase relationships (expressed by
Cochran distribution)
Mlphare
first example of
+The
dm
OAS distribution
solving an unknown
Sim
distribution
protein by directmethod phasing of
the
2.1Å
OAS
data
Solvent flattening
Rusticyanin,
MW: 16.8 kDa; SG: P21;
a=32.43, b=60.68,
c=38.01Å ; b=107.82o ;
Anomalous scatterer: Cu
Oasis
+OAS
dm distribution
Sim distribution
Cochran distribution
Solvent flattening
Radiation damage Induced Phasing
(RIP)
• Radiation damage has been a curse of
macromolecular crystallography from its early
days.
• The X-ray radiation damage of cystals can be
caused by he breakage of covalent bonds as an
immediate consequence of the absorption of an
X-ray quantum (a primary effect) of by the
destructive effect of the propogation of radicals
throughout the crystal (a secondary effect).
• Total dose and dose rate play a role in the
amount of radiation damage inflicted on a
protein crystal.
• The most pronounced structural changes observed were
disulphide-bond breakage and associated main-chain
and side-chain movements as well as decarboxylation of
aspartate and glutamate residues.
• The structural changes induced on the sulphur atoms
were successfully used to obtain high-quality phase
estimates through an RIP (Radiation damage Induced
Phasing) procedure.
Radiation damage Induced Phasing
with Anomalous Scattering (RIPAS)
• Substructure solution and phasing
procedure using a combination of
anomalous scattering and radiation
damage
induced
isomorphous
differences.
• RIPAS strategy is beneficial for both
locating the substructure and subsequent
phasing.
Experimental electron density before solvent flattering with
SAD (left), RIP (middle) and RIPAS (right) phases for the
(a) CS (thaumatin crystal soaked in a diluted
N-iodisuccinamide solution) thaumatin data
(b) IC thaumatin (iodinated crystallized
thaumatin)
Methods of phase improvement
It is not always (!) possible to recognise features in a first electron
density map. There are however ways of improving the map
(phases):
• Solvent Flattening
• Histogram matching
• Non-crystallographic symmetry
(NCS) Averaging
these methods can result in dramatic
improvements in the clarity of the
electron density map.
1. Solvent flattening. Protein crystals contain large amounts of solvent;
this will in general be disordered, and so will not contribute to the crystal
diffraction.
By knowing the protein content of the crystal, it is therefore possible to
determine the threshold density below which is noise; points with
density below the threshold are set to a suitable average value.
This is particularly useful for locating molecular boundaries.
2. Averaging. If the asymmetric unit possesses more than one
molecule, the equivalencing of the various copies can lead to dramatic
improvement in the map and the phases.
Improvement in electron density after solvent
flattening and histogram matching
Before
Green =
solvent envelope
After
Interpretation of the Electron Density
(Building the Model)
• Lots of fun!
• Trace the main-chain
• Try to recognize the
amino acid sequence in
the density.
• Programs:- Xtal view, O
The effect of resolution of the quality of
the electron density map
2.0 Å
1.5 Å
1.2 Å
5.0 Å : see shape of molecule
3.0 Å : see main-chain and some side chains
2.5 Å : see main-chain carbonyls
1.5 Å : ~ atomic resolution.
Resolution
1.2 Å
2Å
3Å
Atomic resolution
Fitting side chains, adding waters
• If the density is good enough you can recognize alternate conformations for side-chains.
• Hydrogens are not seen in the density, except in ultra-high resolutions structures < 1.0 Å.
• Ordered Waters are seen on the surface and occasionally in the interior of the protein.
At 2.0 Å resolution or better ~ 1 water / residue.
Waters molecules play a big role in protein stability and enzyme catalysis.
•Because the density depends on experimental phases which has error associated with them.
The first model can have many errors.
• Therefore it is essential to refine the atomic positions and their thermal parameters.
Chain Tracing
Electron
Density
Chain
Trace
Final
Model
Maps coefficients used
to minimize model bias
2Fo – Fc : most common map seen in paper.
Fo – Fc : (difference map) used with the above map to detect errors
1
( x, y, z )
V
F e
a
2i ( hx ky lz )
hkl
h
k
l
hkl
Refinement Cycle
Refinement: Improving the agreement between the model and the experimental density.
Compare Fobs (From reflection Intensities) to Fcalc (Calculated from the model)
Least squares minimization
Simulated Annealing / Molecular dynamics
Rfactor = numerical indicator to follow progress of refinement
agreement between data and model
data
model
F
F
R
F
obs
Fit Model
calc
obs
data
Calculate map
Refine
Refinement
Refinement
R
# iterations
R = (|Fo-Fc|)/(Fo)
Fc = calculated structure factor
Fo = observed structure factor
The best Fourier is calculated from
1
best(r) =
m|FP(S)|exp(iPbest(S))
S
Protein Data Base growth
Molecular Biology: cloning of genes / over expression of proteins
Synchrotron Radiation: MAD phasing, smaller crystals
Cryo-cooling of crystals: collect data from 1 crystal,
increase order.
Instrumentational and software improvements
Increase in the number of labs using the technique
• Due to the advent of synchrotron radiation and due to
the seleno-methionine derivatization technique, the
total number of protein structures deposited in the
PDB from 1980 onwards has increased
catastrophically.
• MAD technique played a major role in this. At
present nearly 100 new structures are deposited every
week.
THANK YOU