H2N - Department of Computing Science
advertisement

Protein Feature
Identification
David Wishart
Depts. Computing & Biological Science
University of Alberta
david.wishart@ualberta.ca
Proteins
• Exhibit far more sequence and chemical
complexity than DNA or RNA
• Properties and structure are defined by
the sequence and side chains of their
constituent amino acids
• The “engines” of life
• >95% of all drugs target proteins
• Favorite topic of post-genomic era
The Post-genomic Challenge
•
•
•
•
•
•
•
How to rapidly identify a protein?
How to rapidly purify a protein?
How to identify post-trans modification?
How to find information about function?
How to find information about activity?
How to find information about location?
How to find information about structure?
Answer: Look at Protein Features
Protein Features
ACEDFHIKNMF
SDQWWIPANMC
ASDFDPQWERE
LIQNMDKQERT
QATRPQDS...
Sequence View
Structure View
Different Types of Features
• Composition Features
– Mass, pI, Absorptivity, Rg, Volume
• Sequence Features
– Active sites, Binding Sites, Targeting,
Location, Property Profiles, 2o structure
• Structure Features
– Supersecondary Structure, Global Fold,
ASA, Volume
Where To Go
http://www.expasy.org/
Amino Acids (Review)
O
H3N+
H
O
R
Glycine and Proline
H
C
C
H2N
COOH
HN
COOH
H
H
G
P
Aliphatic Amino Acids
CH3
CH3 CH3
CH3
V
H2N
C
COOH
H2N
H
C
I
COOH
H
CH3
CH3
A
H2N
C
H
COOH
CH3
H2N
C
H
COOH
L
Aromatic Amino Acids
N
N
W
H2N
OH
C
COOH
H2N
H
H2N
H
COOH
C
H
Y
C
N
F
H2N
C
H
COOH
H
COOH
Charged Amino Acids
H
N
COO -
D
H2N
C
H2N
COO
C
COOH
NH3+
H
E
H2 N
R
NH
COOH
H
NH3+
K
C
H
COOH
H2N
C
H
COOH
Polar Amino Acids
CONH2
N
H2N
C
COOH
CH3 OH
H2N
H
C
T
COOH
H
CONH2
OH
Q
H2N
S
C
H
COOH
H2N
C
H
COOH
Sulfo-Amino Acids
CH3
S
SH
C
H2N
C
COOH
H2N
COOH
H
H
C
M
Compositional Features
•
•
•
•
•
•
•
Molecular Weight
Amino Acid Frequency
Isoelectric Point
UV Absorptivity
Solubility, Size, Shape
Radius of Gyration
Free Energy of Folding
Molecular Weight
Molecular Weight
•
•
•
•
•
Useful for SDS PAGE and 2D gel analysis
Useful for deciding on SEC matrix
Useful for deciding on MWC for dialysis
Essential in synthetic peptide analysis
Essential in peptide sequencing (classical
or mass-spectrometry based)
• Essential in proteomics and high
throughput protein characterization
Molecular Weight
• Crude MW calculation:
MW = 110 X Numres
• Exact MW calculation:
MW = SAAi x MWi
• Remember to add 1
water (18.01 amu)
after adding all res.
• Note isotopic weights
• Corrections for CHO,
PO4, Acetyl, CONH2
Amino Acid Residue Weights
Residue
A
C
D
E
F
G
H
I
K
L
Weight
71.08
103.14
115.09
129.12
147.18
57.06
137.15
113.17
128.18
113.17
Residue
M
N
P
Q
R
S
T
V
W
Y
Weight
131.21
114.11
97.12
128.14
156.2
87.08
101.11
99.14
186.21
163.18
Amino Acid versus Residue
R
R
C
C
H2N
COOH
H
Amino Acid
N
H
CO
H
Residue
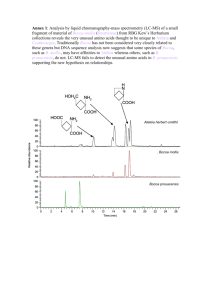
Protein Identification via MW
• MOWSE
• http://srs.hgmp.mrc.ac.uk/cgi-bin/mowse
• CombSearch
• http://ca.expasy.org/tools/CombSearch/
• Mascot
• http://www.matrixscience.com/search_form
_select.html
• AACompSim/AACompIdent
• http://ca.expasy.org/tools/
Molecular Weight & Proteomics
2-D Gel
QTOF Mass Spectrometry
Amino Acid Frequency
• Deviations greater than
2X average indicate
something of interest
• High K or R indicates
possible nucleoprotein
• High C’s indicate stable
but hard-to-fold protein
• High G, P, Q, or N says
lack of stable structure
Table 1
Frequency of amino acid occurrences in water soluble proteins
Residue
A
C
D
E
F
G
H
I
K
L
Frequency
8.80%
2.05%
5.91%
5.89%
3.76%
8.30%
2.15%
5.40%
6.20%
8.09%
Residue
M
N
P
Q
R
S
T
V
W
Y
Frequency
1.97%
4.58%
4.48%
3.84%
4.22%
6.50%
5.91%
7.05%
1.39%
3.52%
Isoelectric Point (pI)
• The pH at which a protein has a net charge=0
•
Q = S Ni/(1 + 10pH-pKi)
Transcendental
equation
pKa Values for Ionizable Amno Acids
Residue
C
D
E
pKa
10.28
3.65
4.25
Residue
H
K
R
pKa
6
10.53
12.43
Isoelectric Point
• Calculation is only approximate (+/- 1 pH)
• Does not include 3o structure interactions
• Can be used in developing purification
protocols via ion exchange chromatography
• Can be used in estimating spot location for
isoelectric focusing gels
• Can be used to decide on best pH to store or
analyze protein
UV Spectroscopy
UV Absorptivity
• UV (Ultraviolet light) has a wavelength of
200 to 400 nm
• Most proteins and peptides (and all nucleic
acids) absorb UV light quite strongly
• UV spectroscopy is the most common form
of spectroscopy performed today
• UV spectra can be used to identify or
classify some proteins or protein classes
UV Absorptivity
• OD280 = (5690 x #W + 1280 x #Y)/MW x Conc.
• Conc. = OD280 x MW/(5690 X #W + 1280 x #Y)
OH
N
H2N
C
H
COOH
H2N
C
H
COOH
Hydrophobicity
• Indicates Solubility
• Indicates Stability
• Indicates Location
(membrane or
cytoplasm)
• Indicates Globularity
or tendency to form
spherical structure
Kyte / Doolittle Hyrophobicity Scale
Residue
A
C
D
E
F
G
H
I
K
L
Hphob
1.8
2.5
-3.5
-3.5
2.8
-0.4
-3.2
4.5
-3.9
3.8
Residue
M
N
P
Q
R
S
T
V
W
Y
Hphob
1.9
-3.5
-1.6
-3.5
-4.5
-0.8
-0.7
4.2
-0.9
-1.3
Hydrophobicity
• Average Hydrophobicity
AH = S AAi x Hi
• Hydrophobic Ratio
RH = S H(-)/S H(+)
• Hydrophobic % Ratio
RHP = %philic/%phobic
• Linear Charge Density
LIND = (K+R+D+E+H+2)/#
• Solubility
SOL = RH + LIND - 0.05AH
• Average AH = 2.5 + 2.5
Insol > 0.1 Unstrc < -6
• Average RH = 1.2 + 0.4
Insol < 0.8 Unstrc > 1.9
• Average RHP = 0.9 + 0.2
Insol < 0.7 Unstrc > 1.4
• Average LIND = 0.25
Insol < 0.2 Unstrc > 0.4
• Average SOL = 1.6 + 0.5
Insol < 1.1 Unstrc > 2.5
Protein Dimensions
• Radius and Radius of Gyration
• Molecular and Partial Specific Volume
• Accessible Surface Area
• Provides a size estimate of a protein
• Used in analytical techniques such as
neutron or X-ray scattering, analytical
ultracentrifugation, light scattering
Radius & Radius of Gyration
• RAD = 3.875 x NUMRES 0.333
(Folded)
• RADG = 0.41 x (110 x NUMRES) 0.5
Radius
(Unfolded)
Radius of Gyration
Partial Specific Volume
• Measured in mL/g
• Inverse measure of
protein density (0.70-75)
• Depends on protein’s
composition and
compactness
• Measured via
sedimentation analysis
• PSV = S PSi x Wi
Table 6
Residue Partial Specific Volumes
Residue
A
C
D
E
F
G
H
I
K
L
PS (ml/g) Residue
0.748
M
0.631
N
0.579
P
0.643
Q
0.774
R
0.632
S
0.67
T
0.884
V
0.789
W
0.884
Y
PS (ml/g)
0.745
0.619
0.774
0.674
0.666
0.613
0.689
0.847
0.734
0.712
Packing Volume
Loose Packing
Dense Packing
Protein
Proteins are Densely Packed
Packing Volume (VP)
• Determined via X-ray
or NMR structure
• “True” measure of
volume occupied by
protein
• Approximate Value
VP = 1.245 x MW
• Exact Value
VP = S AAi x Vi
Table 7
Amino Acid Packing Volumes
3
3
Residue V (Å )
Residue V (Å )
A
88.6
M
162.9
C
108.5
N
117.7
D
111.1
P
122.7
E
138.4
Q
143.9
F
189.9
R
173.4
G
60.1
S
89
H
153.2
T
116.1
I
166.7
V
140
K
168.6
W
227.8
L
166.7
Y
193.6
Different Types of Features
• Composition Features
– Mass, pI, Absorptivity, Rg, Volume
• Sequence Features
– Active sites, Binding Sites, Targeting,
Location, Property Profiles, 2o structure
• Structure Features
– Supersecondary Structure, Global Fold,
ASA, Volume
Sequence Features
AHGQSDFILDEADGMMKSTVPN…
HGFDSAAVLDEADHILQWERTY…
GGGNDEYIVDEADSVIASDFGH…
*[LIVM][LIVM]DEAD*[LIVM][LIVM]*
(EIF 4A ATP DEPENDENT HELICASE)
Probability & Seq. Features
• Expectation value (e) is
the expected number of
hits for a given sequence
pattern or motif
e = N x f1 x f2 x f3 x .... fk
• N is the number of
residues in DB (108)
• fi is the frequency of a
given amino acid(s)
Table 1
Frequency of amino acid occurrences in water soluble proteins
Residue
A
C
D
E
F
G
H
I
K
L
Frequency
8.80%
2.05%
5.91%
5.89%
3.76%
8.30%
2.15%
5.40%
6.20%
8.09%
Residue
M
N
P
Q
R
S
T
V
W
Y
Frequency
1.97%
4.58%
4.48%
3.84%
4.22%
6.50%
5.91%
7.05%
1.39%
3.52%
Example #1
ACIDS
e = 108*0.088*0.021*0.054*0.059*0.065
e = 38.3
#Found in OWL database = 14
Example #2
A*ACI[DEN]S
e = 108*0.088*1.000*0.088*0.021*0.054
*{0.059 + 0.059 + 0.046}*0.065
e = 9.4
#Found in OWL database = 9
Minimum Pattern Lengths
f = 0.08
e = 108*0.088 = 0.17 min = 8
f = 0.05
e = 108*0.057 = 0.08 min = 7
f = 0.03
e = 108*0.036 = 0.07 min = 6
How Long Should a Sequence
Motif or Sequence Block Be?
• How many matching
segments of length “l”
could be found in
comparing a query of
length M to a DB of N ?
• Answer:
n(l) = M x N x fl
• Assume f = 0.05, M =
300, N = 100,000,000
Table 2
n
3,750,000
187,500
9375
469
23
1.2
0.058
l
3
4
5
6
7
8
9
Rule of Thumb
Make your
protein sequence
motifs at least
8 residues long
Sites that Support Pattern
Queries
• OWL Database
– http://bioinf.man.ac.uk/dbbrowser/OWL/
• PIR Website
– http://pir.georgetown.edu/pirwww/search/patmatch.html
• SCNPSITE at EXPASY
– http://ca.expasy.org/tools/scanprosite/
• FPAT (Regular Expression Query)
– http://stateslab.bioinformatics.med.umich.edu/service/fpat/
Regular Expressions
• C[ACG]T - Matches CAT, CCT and CGT only
• C . T - Matches CAT, CaT, C1T, CXT, not CT
• CA?T - Matches CT or CAT only
• C+T - Matches CT, CCT, CCCT, CCCCT…
• C(HE)?A[TP] - Matches CHEAT, CAT, CHEAP, CAP
• S[A-I,L-Q,T-Z]?LK[A-I,L-Q,T-Z]?A - Matches S*LK*A
PROSITE Pattern Expressions
C - [ACG] - T - Matches CAT, CCT and CGT only
C - X -T - Matches CAT, CCT, CDT, CET, etc.
C - {A} -T - Matches every CXT except CAT
C - (1,3) - T - Matches CT, CCT, CCCT
C - A(2) - [TP] - Matches CAAT, CAAP
[LIV] - [VIC] - X(2) - G - [DENQ] - X - [LIVFM] (2) -G
Sequence Feature Databases
• PROSITE - http://ca.expasy.org/prosite/
• BLOCKS - http://www.blocks.fhcrc.org/
• DOMO - http://www.infobiogen.fr/services/domo/
• PFAM - http://pfam.wustl.edu
• PRINTS - http://www.bioinf.man.ac.uk/dbbrowser/PRINTS/
• SEQSITE - PepTool
Phosphorylation Sites
pY
pT
PO4
CH3 PO4
H2N
H2N
pS
C
H
COOH
C
H
PO4
COOH
H2N
C
H
COOH
Phosphorylation Sites
Phopshorylation Sites
>*KRKQI[ST]VR*
CHAN K.F. et al., J. BIOL. CHEM. 257:3655-3659 (1982)
PHOSPHORYLASE KINASE PHOSPHORYLATION SITE
>*KKR**R**[ST]*
KEMP B.E. et al., PNAS 72:3448-3452 (1975)
MYOSIN LIGHT CHAIN KINASE PHOSPHORYLATION SITE
>*NYLRRL[ST]DSNF*
CZERNIK A.J. et al. PNAS 84:7518-7522 (1987)
CALMODULIN DEPENDENT PROTEIN KINASE I PHOSPHORYLATION SITE
Glycosylation
Glycosylation Sites
Glycosylation Sites
>*N!P[ST]!P*
MARSHALL, R.D.W. ANN. REV. BIOCHEM. 41:673-702 (1972)
GLYCOSYLATION SITE (S AND/OR T ARE GLYCOSYLATED)
>*G*K*R*
MARSHALL, R.D.W. ANN. REV. BIOCHEM. 41:673-702 (1972)
GLYCOSYLATION SITE (K IS GLYCOSYLATED)
>*G*K**R*
MARSHALL, R.D.W. ANN. REV. BIOCHEM. 41:673-702 (1972)
GLYCOSYLATION SITE (K IS GLYCOSYLATED)
Signaling
Signaling Sites
Signaling Sites
>*[KRH][DEN]EL$
SMITH M.J. et al., EMBO J. 8:3581-3586 (1989)
ENDOPLASMIC RETICULUM DIRECTING SEQUENCE
>*P***KKRKAV*
KALDERON, D. et al., CELL 39:400-509 (1984)
NUCLEAR TRANSPORT SIGNAL OF SV40 LARGE T ANTIGEN
>${3,20}[LIVFTA][LIVFTA][LIVFTA]{3,6}[LIV]*[GA]C*
VON HEIJNE, G. PROT. ENG. 2:531-534 (1989)
SIGNAL PEPTIDASE II CLEAVAGE SITE
Protease Cut Sites
Protease Cut Sites
Protease Cut Sites
>*[KR]*
*[KR]/*
TRYPSIN CLEAVAGE SITE (CUTS AFTER [KR])
>*[FLY]![VAG]
*/[FLY]![VAG]
PEPSIN CLEAVAGE SITE (CUTS BEFORE [FY])
>*[FWY]*
*[FWY]/*
CHYMOTRYPSIN CLEAVAGE SITE (CUTS AFTER [FWY])
Binding Sites
Binding Sites
>*RGD*
RUOSLAHTII E. et al., CELL 44:517-518 (1986)
FIBRONECTIN ADHESION SITE
>*CDPGYIGSR*
GRAF, J. et al., CELL 48:989-996 (1987)
MAMMAL LAMNIN DOMAIN III B1 CHAIN CELL ATTACHMENT SITE
>*[VIL]**[TS][DN]Y**[FY][AL]*
GODOVAC-ZIMMERMANN, J., TIBS 13:64-66 (1988)
BINDING SITE FOR HYDROPHOBIC MOLECULE TRANSPORT PROTEINS
Family Signature
Sequences
Protein Family Signature Sequences
>*[FY]CRNPD*
NAKAMURA T. et al., NATURE 342:441-445 (1989)
KRINGLE DOMAIN SIGNATURE
>*[LIVM][LIVM]DEAD*[LIVM][LIVM]*
CHANG T.H. et al., PNAS 87:1571-1575 (1990)
EIF 4A FAMILY ATP DEPENDENT HELICASE SIGNATURE
>*C*C*****G**C*
BLOMQUIST M.C. et al., PNAS 81:7363-7362 (1984)
EGF/TGF SIGNATURE SEQUENCE
Enzyme Active Sites
Enzyme Active Sites
>*[MAFILV]DTG[STA][STAN]*
DOOLITTLE, R.F., OF URFS AND ORFS, 1986
ACID OR ASPARTYL PROTEASE ACTIVE SITE
>*TCP&NLGT*
DOOLITTLE, R.F., OF URFS AND ORFS, 1986
GUANIDINE KINASE ACTIVE SITE
>*F*[LIVFMY]*S**K****[AG]*[LIVM]L*
JORIS, B. ET AL., BIOCHEM. J. 250:313-324 (1989)
BETA LACTAMASE (TYPE A) ACTIVE SITE
T-Cell Epitopes
•
•
•
•
Type I peptides are 8 - 10 amino acids
Type II peptides are 12 - 20 amino acids
Type I are endogenous, Type II exogenous
Suggestion they are amphipathic helices
• HLA-A1
*[ED]P****[YF]
• A2.1
***[AVILF][AVILF][AVILF]***
• HLA-DR1b[YF]**[ML]*[GA]**L
Better Methods for
Sequence Feature ID
• Sequence Profiles/Scoring Matrices
• Neural Networks
• Hidden Markov Models
• Bayesian Belief Nets
• Reference Point Logistics
A Sample Sequence Profile
A
C
D
E
F
G
1 W G V L
V 3 -2
3
4
0
2 L L S P
L 2 -2 -2 -1
3 V V V V
V 2
4 K E A T
A 6 -2
5
6 -5
4
1
0
5 A P L P
P 6 -1
0
1 -2
2
0
1
6 G G G G
G 7
1
7
7 S S Q E
D 4 -1
8 S S T P
S 4
I
K
L
M
N
P
Q
R
S
T
V
4 -1
3 -1
4
4
1
1
1 -2
1
2
6 -6 -2
3
0 -1
3 -1
6
5 -1
3
0 -1
3
1
4
2
2 -3 11 -2
8
6 -2
1 -2 -2
0
2 15 -9 -1
5 -2
0
3
3
3
1
3
6
0 -6 -4
0
2
0
8
2
0
2
2
3 -5 -4
5 -6 15 -1 -3
0 -4 -3
4
3
6
1
6
2 -1 -6 -5
7
7 -6
7
2 -3 -2
4
3
6
1
6
2 -1 -6 -5
2
2 -4
4 -1
2 -3 -2
2
7
0
1 10
2 -2 -2
4
H
2 -2
0
2
<e>i = log2(qi/pi)
6
W
Y
1 -1
0 -2 -4
Calculating a Profile Score
A
C
D
E
F
G
K
L
M
N
P
Q
R
S
T
V
1 W G V L
V 3 -2
3
4
0
4 -1
3 -1
4
4
1
1
1 -2
1
2
6 -6 -2
2 L L S P
L 2 -2 -2 -1
3
0 -1
3 -1
6
5 -1
3
0 -1
3
1
4
3 V V V V
V 2
2
2 -3 11 -2
8
6 -2
1 -2 -2
0
2 15 -9 -1
4 K E A T
A 6 -2
5
6 -5
4
1
0
5 -2
0
3
3
3
1
3
6
0 -6 -4
5 A P L P
P 6 -1
0
1 -2
2
0
1
0
2
0
8
2
0
2
2
3 -5 -4
6 G G G G
G 7
1
7
5 -6 15 -1 -3
0 -4 -3
4
3
6
1
6
2 -1 -6 -5
7 S S Q E
D 4 -1
7
7 -6
7
2 -3 -2
4
3
6
1
6
2 -1 -6 -5
8 S S T P
S 4
2
2 -4
4 -1
2 -3 -2
2
7
0
1 10
2 -2 -2
4
H
I
2 -2
0
2
6
W
Y
1 -1
0 -2 -4
VLVAPGDS = 6+6+15+6+8+15+7+10=66
LVLGPGLA = 4+4+8+4+8+15-3+4= 44
Hidden Markov Models
Neural Networks
nodes
Training
Set
Layer 1 Hidden
Layer
Output
What Can Be Predicted?
•
•
•
•
•
•
•
•
•
O-Glycosylation Sites
Phosphorylation Sites
Protease Cut Sites
Nuclear Targeting Sites
Mitochondrial Targ Sites
Chloroplast Targ Sites
Signal Sequences
Signal Sequence Cleav.
Peroxisome Targ Sites
•
•
•
•
•
•
•
•
•
ER Targeting Sites
Transmembrane Sites
Tyrosine Sulfation Sites
GPInositol Anchor Sites
PEST sites
Coil-Coil Sites
T-Cell/MHC Epitopes
Protein Lifetime
A whole lot more….
Cutting Edge Sequence
Feature Servers
• Membrane Helix Prediction
– http://www.cbs.dtu.dk/services/TMHMM-2.0/
• T-Cell Epitope Prediction
– http://syfpeithi.bmiheidelberg.com/scripts/MHCServer.dll/home.htm
• O-Glycosylation Prediction
– http://www.cbs.dtu.dk/services/NetOGlyc/
• Phosphorylation Prediction
– http://www.cbs.dtu.dk/services/NetPhos/
• Protein Localization Prediction
– http://psort.nibb.ac.jp/
Subcellular Localization
http://www.cs.ualberta.ca/~bioinfo/PA/Sub/
Profiles & Motifs are Useful
• Helped identify active site of HIV protease
• Helped identify SH2/SH3 class of STP’s
• Helped identify important GTP oncoproteins
• Helped identify hidden leucine zipper in HGA
• Used to scan for lectin binding domains
• Regularly used to predict T-cell epitopes
Score
Amino Acid Property Profiles
3
2
1
0
-1
-2
-3
-4
1
51
101
151
201
251
301
Amino Acid Property Profiles
• Intent is to predict protein’s physical
properties directly from sequence as
opposed to composition or wet chemistry
• Offers a more detailed, graphical view of
sequence-specific properties than
compositional analysis (more powerful?)
• Underlying assumption is: amino acid
properties are additive
Property Profile Algorithm
• Assign each residue a numeric value
corresponding to the physical property
• Choose an odd numbered window (5 or 7)
and calculate the average value
• Assign the average value to the middle
residue in the window
• Move the window down by one residue and
repeat steps 1 to 4 until finished - PLOT
Common Property Profiles
• Hydrophobicity (Watch Scales!)
• Helical Wheel (Not a True Profile)
• Hydrophobic Moments (Helix & Beta sheet)
• Flexibility (Thermal B Factors)
• Surface Accessibility (ASA)
• Antigenicity (B-cell epitopes/T-cell epitopes)
Hydrophobicity Profile
• Plotted using: <H>i = S Hn/(2k + 1)
• Shows location of membrane spanning
regions, epitopes, surface exposed AA’s, etc.
Helical Wheel
• Used to identify disposition of AA side
chains around a helix, looking end-on
• Identifies Helical Amphipathicity
Hydrophobic Moment
• Quantitative way to measure amphipathicity
• Fourier Transform of hydrophobicity
H = {[SHnsin(n)]2 + [SHncos(n)]2}1/2
Flexibility
Flexibility (A^2)
• B factors from X-ray crystallography
• Potentially identifies antigenic and active
sites from sequence data alone
12
11.5
11
10.5
10
9.5
9
8.5
8
1
11
21
31
41
51
61
71
81
91
101
Membrane Spanning Regions
Predicting via Hydrophobicity
Bacteriorhodopsin
4
2
OmpA
3
1.5
2
1
0.5
1
0
0
-0.5
1
-1
-1
-2
-1.5
-3
-2
Bacteriorhodoposin
OmpA
Predicting via Hydrophobicity
Quality of Membrane Helix Prediction of Membrane Proteins.
Protein
Technique Predicted #helices
Actual #helices
Engelman et al.
10
Microsomal cytochrome Chou & Fasman
8
1
p 450
Rao & Argos
5
AMP07
1
Eisenberg et al.
8
Kyte-Doolittle
5
Fo-F1 ATPase (subunit A)
4
Rao & Argos
4
AMP07
4
Jahnig
6
Eisenberg et al.
1
Photosynthetic Reaction Rose
4
5
Centre (M chain)
Kyte-Doolittle
4
Klein et al.
5
Jahnig
7
Kyte-Doolittle
4
Bacteriorhodopsin
67
Engelman et al.
7
Klein et al.
7
Predicting via Neural Nets
• PHDhtm
http://cubic.bioc.columbia.edu/predictprotein/submit_adv.html
• TMAP
http://www.mbb.ki.se/tmap/index.html
• TMPred
http://www.ch.embnet.org/software/TMPRED_form.html
ACDEGF...
Prediction Performance
Secondary Structure Prediction
Secondary Structure Prediction
•
•
•
•
•
•
•
Statistical (Chou-Fasman, GOR)
Homology or Nearest Neighbor (Levin)
Physico-Chemical (Lim, Eisenberg)
Pattern Matching (Cohen, Rooman)
Neural Nets (Qian & Sejnowski, Karplus)
Evolutionary Methods (Barton, Niemann)
Combined Approaches (Rost, Levin, Argos)
Chou-Fasman Statistics
Table 8
Chou & Fasman Secondary Structure Propensity of the Amino Acids
A
C
D
E
F
G
H
I
K
L
Pa
1.42
0.7
1.01
1.51
1.13
0.57
1
1.08
1.16
1.21
Pb
0.83
1.19
0.54
0.37
1.38
0.75
0.87
1.6
0.74
1.3
Pc
0.75
1.11
1.45
1.12
0.49
1.68
1.13
0.32
1.1
0.49
M
N
P
Q
R
S
T
V
W
Y
Pa
1.45
0.67
0.57
1.11
0.98
0.77
0.83
1.06
1.08
0.69
Pb
1.05
0.89
0.55
1.1
0.93
0.75
1.19
1.7
1.37
1.47
Pc
0.5
1.44
1.88
0.79
1.09
1.48
0.98
0.24
0.45
0.84
The PhD Approach
PRFILE...
PHD
ZHANG
GOR III
JASEP7
PTIT
LEVIN
LIM
GOR I
CF
Scores (%)
Prediction Performance
75
70
65
60
55
50
45
Best of the Best
• PredictProtein-PHD (72%)
– http://cubic.bioc.columbia.edu/predictprotein
• Jpred (73-75%)
– http://www.compbio.dundee.ac.uk/~www-jpred/
• SABLE (75%)
– http://sable.chmcc.org/
• PSIpred (77%)
– http://bioinf.cs.ucl.ac.uk/psipred/
• Proteus (78-90%)
– http://wishart.biology.ualberta.ca/proteus/index.shtml
The Proteus Server
EVA- http://cubic.bioc.columbia.edu/eva/
Different Types of Features
• Composition Features
– Mass, pI, Absorptivity, Rg, Volume
• Sequence Features
– Active sites, Binding Sites, Targeting,
Location, Property Profiles, 2o structure
• Structure Features
– Supersecondary Structure, Global Fold,
ASA, Volume
3D Protein Features
Secondary Structure
Table 10
Phi & Psi angles for Regular Secondary
Structure Conformations
Structure
Antiparallel b-sheet
Parallel b-Sheet
Right-handed a-helix
310 helix
p helix
Polyproline I
Polyproline II
Polyglycine II
Phi (F)
-139
-119
-+64
-49
-57
-83
-78
-80
Psi(Y)
+135
+113
+40
-26
-70
+158
+149
+150
Supersecondary Structure
Global Folds
Lactate
Dehydrogenase:
Mixed a / b
Immunoglobulin
Fold: b
Hemoglobin B
Chain: a
3D Structure
• Allows direct identification and/or
location of cofactors, ligands,
crevices, protrusions and other
features
• Allows one to identify possible
function (through 3D homology)
• Allows protein to be classified into a
folding family
3D Structure Classifiers
• CATH
– http://www.biochem.ucl.ac.uk/bsm/cath/
• VAST
– http://www.ncbi.nlm.nih.gov/Structure/VAST/va
stsearch.html/
• Combinatorial Extension (CE)
– http://cl.sdsc.edu/ce.html
• FSSP/Dali
– http://www.ebi.ac.uk/dali/Interactive.html
Accessible Surface Area
Accessible Surface Area
Reentrant Surface
Solvent Probe
Accessible Surface
Van der Waals Surface
ASA -- A Powerful Tool
• Provides a picture of how water or
other small molecules “see” the
protein
• Allows identification of exterior
features from interior features
• Allows identification of protrusions
or crevices (i.e. active sites or
binding sites)
Surface Charge Distribution
Surface Charge
• Allows positively and negatively
charged structural features
(protrusions, crevices) to be
identified
• Can be used to ID possible active
sites or probably character of ligands
• Key to many drug design efforts
Structure Features
•
•
•
•
•
•
•
•
•
Secondary Structure
Supersecondary Structure
Folding Class
Polar/Nonpolar ASA
Hydrogen Bond Parameters
Stereochemistry
Packing Defects
Surface Charge Distribution
Surface Roughness
http://redpoll.pharmacy.ualberta.ca
Conclusion
• Composition Features
– Mass, pI, Absorptivity, Rg, Volume
• Sequence Features
– Active sites, Binding Sites, Targeting,
Location, Property Profiles, 2o structure
• Structure Features
– Supersecondary Structure, Global Fold,
ASA, Volume