DICTA2012-On Computing Greyscale Morphology - spat-var
On Computing Greyscale Morphology with Large Exact Spheres in Arbitrary Dimensions
Via 1-D Distance Transforms
Richard Beare
Department Of Medicine, Monash University &
Developmental Imaging, Murdoch Children’s Research Institute
Paul Jackway
CSIRO Mathematics, Informatics and Statistics,
CSIRO Transformational Biology Capability Platform
MATHEMATICS, INFORMATICS AND STATISTICS
We present a novel constant time algorithm for greyscale (hyper-)spherical flat dilations and erosions. The method is: exactly isotropic, time-independent of the structuring function size, and inherently parallelizable at several levels of granularity. Subsequent different size dilations (or erosions) of the same image may also be performed at insignificant further cost.
Morphology With Large Structuring Elements
Many applications of Mathematical Morphology require large isotropic
Structuring Elements (SE). This presents a problem: large digital circles or spheres contain O(r k ) points, where r is the radius and k the dimensionality, and do not decompose in any of the ways which makes the well-known computational speed-ups of morphology possible. We therefore expect the computational cost of a naive implementation to scale as O(r k ) which quickly becomes prohibitive for large r and high k.
Well-known polygonal approximations to circles and spheres, although separable into compositions of fast linear (1-D) morphology, quickly become obviously non-isotropic at larger r and this effect is worse in higher dimensions. They also do not allow precise control of SE radius.
We present here an exact method based on thresholds of the squared
Euclidean distance transform (EDT 2 ) which we can efficiently compute in
k-dimensions as a cascade of k, one-dimensional EDT
2 s.
Comparative Performance
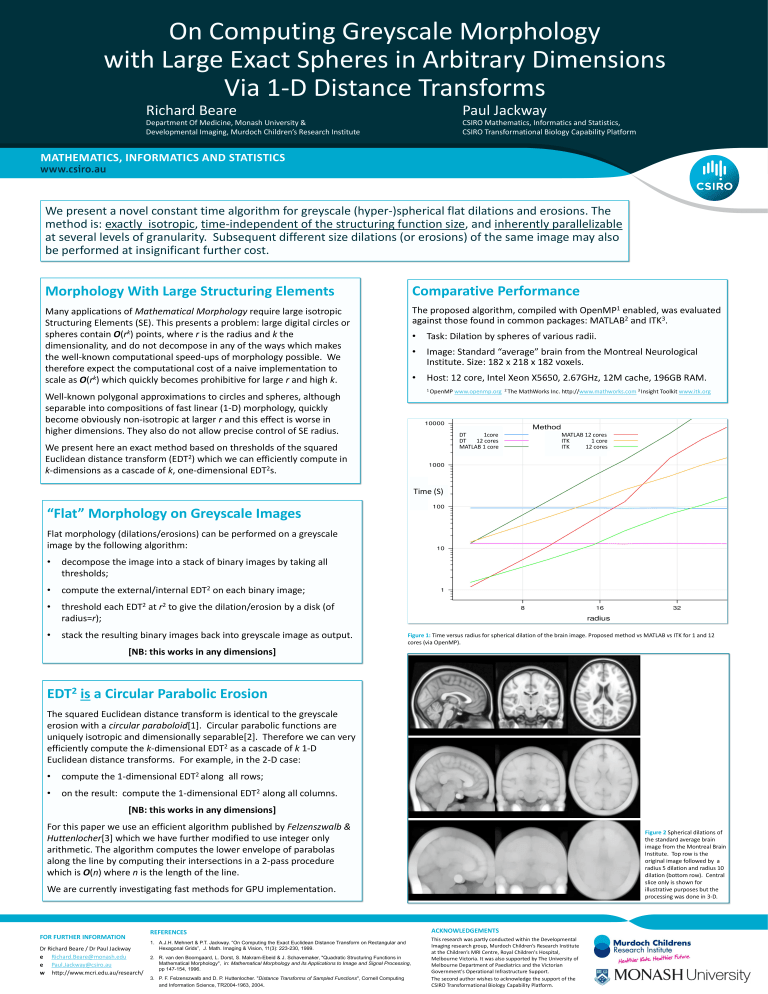
The proposed algorithm, compiled with OpenMP 1 enabled, was evaluated against those found in common packages: MATLAB 2 and ITK 3 .
• Task: Dilation by spheres of various radii.
• Image: Standard “average” brain from the Montreal Neurological
Institute. Size: 182 x 218 x 182 voxels.
• Host: 12 core, Intel Xeon X5650, 2.67GHz, 12M cache, 196GB RAM.
1 OpenMP www.openmp.org
2 The MathWorks Inc. http:// www.mathworks.com
3 Insight Toolkit www.itk.org
DT 1core
DT 12 cores
MATLAB 1 core
MATLAB 12 cores
ITK 1 core
ITK 12 cores
Time (S)
“Flat” Morphology on Greyscale Images
Flat morphology (dilations/erosions) can be performed on a greyscale image by the following algorithm:
• decompose the image into a stack of binary images by taking all thresholds;
• compute the external/internal EDT 2 on each binary image;
• threshold each EDT 2 at r 2 to give the dilation/erosion by a disk (of radius=r);
• stack the resulting binary images back into greyscale image as output.
[NB: this works in any dimensions]
Figure 1: Time versus radius for spherical dilation of the brain image. Proposed method vs MATLAB vs ITK for 1 and 12 cores (via OpenMP).
EDT
2
is a Circular Parabolic Erosion
The squared Euclidean distance transform is identical to the greyscale erosion with a circular paraboloid[1]. Circular parabolic functions are uniquely isotropic and dimensionally separable[2]. Therefore we can very efficiently compute the k-dimensional EDT 2 as a cascade of k 1-D
Euclidean distance transforms. For example, in the 2-D case:
• compute the 1-dimensional EDT 2 along all rows;
• on the result: compute the 1-dimensional EDT 2 along all columns.
[NB: this works in any dimensions]
For this paper we use an efficient algorithm published by Felzenszwalb &
Huttenlocher[3] which we have further modified to use integer only
arithmetic. The algorithm computes the lower envelope of parabolas along the line by computing their intersections in a 2-pass procedure which is O(n) where n is the length of the line.
We are currently investigating fast methods for GPU implementation.
Figure 2 Spherical dilations of the standard average brain image from the Montreal Brain
Institute. Top row is the original image followed by a radius 5 dilation and radius 10 dilation (bottom row). Central slice only is shown for illustrative purposes but the processing was done in 3-D.
FOR FURTHER INFORMATION
Dr Richard Beare / Dr Paul Jackway e Richard.Beare@monash.edu
e Paul.Jackway@csiro.au
w http://www.mcri.edu.au/research/
REFERENCES
1. A.J.H. Mehnert & P.T. Jackway. “On Computing the Exact Euclidean Distance Transform on Rectangular and
Hexagonal Grids”, J. Math. Imaging & Vision, 11(3): 223-230, 1999.
2. R. van den Boomgaard, L. Dorst, S. Makram-Ebeid & J. Schavemaker, "Quadratic Structuring Functions in
Mathematical Morphology", in: Mathematical Morphology and its Applications to Image and Signal Processing , pp 147-154, 1996.
3. P. F. Felzenszwalb and D. P. Huttenlocher. " Distance Transforms of Sampled Functions ", Cornell Computing and Information Science, TR2004-1963, 2004 .
ACKNOWLEDGEMENTS
This research was partly conducted within the Developmental
Imaging research group, Murdoch Children’s Research Institute at the Children’s MRI Centre, Royal Children's Hospital,
Melbourne Victoria. It was also supported by The University of
Melbourne Department of Paediatrics and the Victorian
Government's Operational Infrastructure Support.
The second author wishes to acknowledge the support of the
CSIRO Transformational Biology Capability Platform.