Feb 23

Prepare a 10’talk for Friday Feb 27 on plant defense responses or describe interactions between plants & pathogens or symbionts
Plant defense responses
• Hypersensitive response
• Systemic acquired resistance
• Innate immunity
• Phytoalexin synthesis
• Defensins and other proteins
• Oxidative burst
Some possible pathogens
• Agrobacterium tumefaciens
• Agrobacterium rhizogenes
• Pseudomonas syringeae
• Pseudomonas aeruginosa
• Viroids
• DNA viruses
• RNA viruses
• Fungi
• Oomycetes
Some possible symbionts
• N-fixing bacteria
• N-fixing cyanobacteria
• Endomycorrhizae
• Ectomycorrhizae
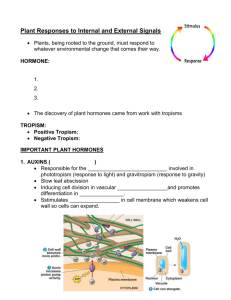
1.Auxins
2.Cytokinins
3.Gibberellins
4.Abscisic Acid
5.Ethylene
6.Brassinoteroids
7.Jasmonic Acid
8.Salicylic Acid
9.Strigolactones
10.Nitric Oxide
11.Sugars
Growth regulators
Auxin signaling
Auxin receptors eg TIR1 are E3 ubiquitin ligases !
Upon binding auxin they activate complexes targeting AUX/IAA proteins for degradation!
AUX/IAA inhibit ARF transcription factors, so this turns on
"early genes"
Some early genes turn on
'late genes" needed for development
Auxin signaling
ABP1 is a different IAA receptor localized in ER
• Activates PM H+ pump by sending it to PM & keeping it there
• Does not affect gene expression!
Auxin & other growth regulators
• Some "late genes" synthesize ethylene (normally a wounding response): how 2,4-D kills?
• Auxin/cytokinin determines whether callus forms roots or shoots
Cytokinins
Discovered as factors which induce cultured cells to divide
Haberlandt (1913): phloem chemical stimulates division
Cytokinins
Discovered as factors which induce cultured cells to divide
Haberlandt (1913): phloem chemical stimulates division van Overbeek (1941): coconut milk stimulates division
Cytokinins
Discovered as factors which induce cultured cells to divide
Haberlandt (1913): phloem chemical stimulates division van Overbeek (1941): coconut milk stimulates division
Miller… Skoog (1955): degraded DNA stimulates division!
Cytokinins
Discovered as factors which induce cultured cells to divide
Haberlandt (1913): phloem chemical stimulates division van Overbeek (1941): coconut milk stimulates division
Miller… Skoog (1955): degraded DNA stimulates division!
Kinetin was the breakdown product
Cytokinins
Discovered as factors which induce cultured cells to divide
Haberlandt (1913): phloem chemical stimulates division van Overbeek (1941): coconut milk stimulates division
Miller… Skoog (1955): degraded DNA stimulates division!
Kinetin was the breakdown product
Derived from adenine
Cytokinins
Discovered as factors which induce cultured cells to divide
Haberlandt (1913): phloem chemical stimulates division van Overbeek (1941): coconut milk stimulates division
Miller… Skoog (1955): degraded DNA stimulates division!
Kinetin was the breakdown product
Derived from adenine
Requires auxin to stimulate division
Cytokinins
Requires auxin to stimulate division
Kinetin/auxin determines tissue formed (original fig)
Cytokinins
Requires auxin to stimulate division
Kinetin/auxin determines tissue formed
Inspired search for natural cytokinins
Miller& Letham (1961) ± simultaneously found zeatin in corn
Kinetin transZeatin
Cytokinins
Miller& Letham (1961) ± simultaneously found zeatin
Later found in many spp including coconut milk
Kinetin transZeatin
Cytokinins
Miller& Letham (1961) ± simultaneously found zeatin
Later found in many spp including coconut milk
Trans form is more active, but both exist (& work)
Many other natural & synthetics have been identified
Cytokinins
Many other natural & synthetics have been identified
Like auxins, many are bound to sugars or nucleotides
Cytokinins
Many other natural & synthetics have been identified
Like auxins, many are bound to sugars or nucleotides
Inactive, but easily converted
Cytokinin Synthesis
Most cytokinins are made at root apical meristem & transported to sinks in xylem
Cytokinin Synthesis
Most cytokinins are made at root apical meristem & transported to sinks in xylem
Therefore have inverse gradient with IAA
Cytokinin Synthesis
Most cytokinins are made at root apical meristem & transported to sinks in xylem
Therefore have inverse gradient with IAA
Why IAA/CK affects development
Cytokinin Synthesis
Most cytokinins are made at root apical meristem & transported to sinks in xylem
Therefore have inverse gradient with IAA
Why IAA/CK affects development
Rapidly metabolized by sink
Cytokinin Effects
Regulate cell division
•
Need mutants defective in CK metabolism or signaling to detect this in vivo
Cytokinin Effects
Regulate cell division
•
Need mutants defective in CK metabolism or signaling to detect this in vivo
• SAM & plants are smaller when
[CK]
Cytokinin Effects
•
SAM & plants are smaller when [CK]
•
Roots are longer!
Cytokinin Effects
•
Usually roots have too much CK: inhibits division!
•
Cytokinins mainly act @ root & shoot meristems
Cytokinin Effects
Cytokinins mainly act @ root & shoot meristems
Control G1-> S & G2-> M transition
Cytokinin Effects
•
Promote lateral bud growth
Cytokinin Effects
•
Promote lateral bud growth
•
Delay leaf senescence
Cytokinin Effects
•
Promote lateral bud growth
•
Delay leaf senescence
• Promote cp development, even in dark
Cytokinin Receptors
Receptors were identified by mutation
Resemble bacterial 2-component signaling systems
Cytokinin Action
1.Cytokinin binds receptor's extracellular domain
Cytokinin Action
1.Cytokinin binds receptor's extracellular domain
2. Activated protein kinases His kinase & receiver domains
Cytokinin Action
1.Cytokinin binds receptor's extracellular domain
2. Activated protein kinases His kinase & receiver domains
3. Receiver kinases His-P transfer relay protein (AHP)
Cytokinin Action
1.Cytokinin binds receptor's extracellular domain
2. Activated protein kinases His kinase & receiver domains
3. Receiver kinases His-P transfer relay protein (AHP)
4. AHP-P enters nucleus & kinases ARR response regulators
Cytokinin Action
4. AHP-P enters nucleus & kinases ARR response regulators
5. Type B ARR induce type A
Cytokinin Action
4. AHP-P enters nucleus & kinases ARR response regulators
5. Type B ARR induce type A
6. Type A create cytokinin responses
Cytokinin Action
4. AHP-P enters nucleus & kinases ARR response regulators
5. Type B ARR induce type A
6. Type A create cytokinin responses
7. Most other effectors are unknown but D cyclins is one effect.
Auxin & other growth regulators
• Some "late genes" synthesize ethylene (normally a wounding response): how 2,4-D kills?
• Auxin/cytokinin determines whether callus forms roots or shoots
• Auxin induces Gibberellins
Gibberellins
Discovered by studying "foolish seedling" disease in rice
• Hori (1898): caused by a fungus
Gibberellins
Discovered by studying "foolish seedling" disease in rice
• Hori (1898): caused by a fungus
• Sawada (1912): growth is caused by fungal stimulus
Gibberellins
Discovered by studying "foolish seedling" disease in rice
• Hori (1898): caused by a fungus
• Sawada (1912): growth is caused by fungal stimulus
• Kurosawa (1926): fungal filtrate causes these effects
Gibberellins
Discovered by studying "foolish seedling" disease in rice
• Kurosawa (1926): fungal filtrate causes these effects
• Yabuta (1935): purified gibberellins from filtrates of
Gibberella fujikuroi cultures
Gibberellins
Discovered by studying "foolish seedling" disease in rice
• Kurosawa (1926): fungal filtrate causes these effects
• Yabuta (1935): purified gibberellins from filtrates of
Gibberella fujikuroi cultures
• Discovered in plants in 1950s
Discovered in plants in 1950s
Gibberellins
• "rescued" some dwarf corn & pea mutants
Discovered in plants in 1950s
Gibberellins
• "rescued" some dwarf corn & pea mutants
• Made rosette plants bolt
Discovered in plants in 1950s
Gibberellins
• "rescued" some dwarf corn & pea mutants
• Made rosette plants bolt
• Trigger adulthood in ivy & conifers
Gibberellins
• "rescued" some dwarf corn & pea mutants
• Made rosette plants bolt
• Trigger adulthood in ivy & conifers
• Induce growth of seedless fruit
Gibberellins
•
"rescued" some dwarf corn & pea mutants
•
Made rosette plants bolt
• Trigger adulthood in ivy & conifers
• Induce growth of seedless fruit
•
Promote seed germination
Gibberellins
•
"rescued" some dwarf corn & pea mutants
•
Made rosette plants bolt
• Trigger adulthood in ivy & conifers
• Induce growth of seedless fruit
•
Promote seed germination
•
Inhibitors shorten stems: prevent lodging
Gibberellins
•
"rescued" some dwarf corn
& pea mutants
• Made rosette plants bolt
• Trigger adulthood in ivy
& conifers
•
Induce growth of seedless fruit
• Promote seed germination
• Inhibitors shorten stems: prevent lodging
•
>136 gibberellins (based on structure)!
Gibberellins
>136 gibberellins (based on structure)!
• Most plants have >10
Gibberellins
>136 gibberellins (based on structure)!
• Most plants have >10
• Activity varies dramatically!
Gibberellins
>136 gibberellins (based on structure)!
• Most plants have >10
• Activity varies dramatically!
•
Most are precursors or degradation products
Gibberellins
>136 gibberellins (based on structure)!
• Most plants have >10
• Activity varies dramatically!
• Most are precursors or degradation products
• GAs 1, 3 & 4 are most bioactive
Gibberellin signaling
Used mutants to learn about GA signaling
Gibberellin signaling
Used mutants to learn about GA signaling
•
Many are involved in GA synthesis
Gibberellin signaling
Used mutants to learn about GA signaling
•
Many are involved in GA synthesis
•
Varies during development
Gibberellin signaling
Used mutants to learn about GA signaling
•
Many are involved in GA synthesis
•
Varies during development
•
Others hit GA signaling
•
Gid = GA insensitive
Gibberellin signaling
Used mutants to learn about GA signaling
•
Many are involved in GA synthesis
•
Varies during development
•
Others hit GA signaling
•
Gid = GA insensitive
• encode GA receptors
Gibberellin signaling
Used mutants to learn about GA signaling
•
Many are involved in GA synthesis
•
Varies during development
•
Others hit GA signaling
•
Gid = GA insensitive
• encode GA receptors
•
Sly = E3 receptors
Gibberellin signaling
Used mutants to learn about GA signaling
•
Many are involved in GA synthesis
•
Varies during development
•
Others hit GA signaling
•
Gid = GA insensitive
• encode GA receptors
•
Sly = E3 receptors
•
DELLA (eg rga) = repressors of GA signaling
Gibberellins
GAs 1, 3 & 4 are most bioactive
Act by triggering degradation of DELLA repressors
Gibberellins
GAs 1, 3 & 4 are most bioactive
Made at many locations in plant
Act by triggering degradation of DELLA repressors w/o GA DELLA binds & blocks activator (GRAS)
Gibberellins
Act by triggering degradation of DELLA repressors w/o GA DELLA binds & blocks activator bioactive GA binds GID1; GA-GID1 binds DELLA & marks for destruction
Gibberellins
Act by triggering degradation of DELLA repressors w/o GA DELLA binds & blocks activator bioactive GA binds GID1; GA-GID1 binds DELLA & marks for destruction
GA early genes are transcribed, start
GA responses