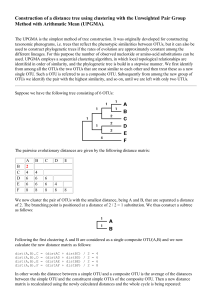
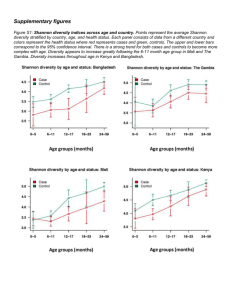
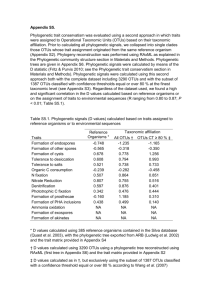
4_OTUS_OPUS_SPECIES
advertisement

Species OTUs OPUs Species OTUs OPUs SPECIES CONCEPT versus DEFINITION The CONCEPT is the IDEA, what embraces a unit (generally immutable) The DEFINITION is the WAY to embrace a unit (changes with technical developments) (depends on the observable characters) Taxa circumscription depends on the observable characters Most plants & animals Rest of eukaryotes Prokaryotes Sexual isolation (biological SC) Morphology (taxonomic SC) Genealogy / Genetics / Phenotype Rosselló-Mora & Amann 2001, FEMS Rev. 25:39-67 CONCEPT vs DEFINITION The CONCEPT is the IDEA, what embraces a unit (generally immutable) monophyletic group of isolates genomically coherent sharing high similarity in many independent phenotypic features The DEFINITION is the WAY to embrace a unit (changes with technical developments) monophyly gene sequence analysis (i.e. 16S rRNA) genomic coherence DDH phenotype (biochemical tests, chemotaxonomy…) How do we define / circumscribe species for prokaryotes phylogenetic coherence genomic coherence phenotypic coherence 50% 60% 70% 70-50% 70% 80% 100% RNAr 16S Functional genes (MLSA) Genomic analyses Reasociación DNA-DNA G+C, AFLP, MLSA Genomic comparisons (ANI; AAI) metabolism chemotaxonomy spectrometry (Maldi-Tof; ICR-FT/MS) 16S rRNA gene sequence (gold sdt) DDH (gold standard) identificative phenotypic property 97% - 98.7% identity threshold 70% similarity threshold chemotaxonomic markers all organisms must be monophyletic 96% ANI metabolic homogeneity Tindall et al., 2010 IJSEM 60:249-266 The problems of having to isolate organisms for taxonomic studies: it is impossible to “formally” classify uncultured organisms yet just the Candidatus status is recognized difficulties to homogenize names for the scientific community The benefits of having to isolate organisms for taxonomic studies: validation of a name requires DEPOSIT of a type strain in TWO international collections type material should be AVAILABLE to all the scientific community type strains are the REFERENCE for any taxonomic work Species microbial molecular ecology MOLECULAR TECHNIQUES generally identification of units (species) by means of 16S rRNA genes generally inform about the highly abundant organisms it is not clear where to set a threshold of what is a species Red => knowable diversity / black => seed bank, unknown, difficult to know Pedrós-Alió, 2006 TRENDS Microbiol 14:257-263 ≠ disciplines use ≠ size of their basic units ≠ observational methods Taxonomists phylogenetic / genomic / phenotypic coherence Ecologists 97% identity OTUs too wide for taxonomists Evolutionary microbiologists much more strict too narrow for taxonomists Compile microdiversity Ecotype; early stage of speciation A stable framework needs PRAGMATISM into OTUs at 97% identity Rosselló-Móra, 2011 Environ Microbiol 14:318-334 OTUs OPERATIONAL TAXONOMIC UNITS Clustering by sequence identity threshold Clustering at XX% identity Quiime … Metagenome 5’ V1 & V2 OTU 1 V5 & V6 OTU 2 OTU 3 OTU 4 Singletons doubletons 3’ Different groups use different variable zones metagenomes of different zones are not comparable perhaps identical sequences of different stretches may match different OTUs (green highlighted) High identity does not mean common ancestry OTUs OPERATIONAL TAXONOMIC UNITS 97% sequence identity threshold 100% 100% reconditioning 99% 98% 97% Clone libraries great phylotype diversity PCR errors (reconditioning) microdiversity (several operons?) grouping through % identity OTU (Operational Taxonomic Unit) 97% one species? Acinas et al., 2004 Nature 430:551-554 RECOMMENDATIONS FOR THE USE OF OTUS Use 99% or 98.7% identity (IS AT THE RESOLUTION OF A TAXONOMIC SPECIES) Ecologists 97% identity OTUs too wide for taxonomists Evolutionary microbiologists much more strict too narrow for taxonomists 450000 SILVA REF 112 DATABASE 400000 SEQUENCING ERRORS 350000 300000 250000 200000 150000 99,50% 100000 99,00% 98,70% 50000 0 0 300000 600000 900000 1200000 Yarza et al., Nature Revs. 2014. 12: 635-645 OPUs OPERATIONAL PHYLOGENETIC UNITS Grouping after phylogenetic inference (using parsimony tool of ARB) Clustering at XX% identity Quiime … Metagenome OTU 1 OTU 2 OTU 3 (representatives) OTU 4 Singletons doubletons Selected sequences are inserted in a tree preexisting using parsimony inference different sizes, zones, may affiliate together if no use of ARB, one can select the best sequences from the databases matching OTUs and reconstruct properly One OPU will contain different OTUs of different length, zone, sample, etc… OPUs subjective best solution to measure diversity OPU Operational Phylogenetic Unit similar to “Operational Phylogenetic-based Microbial Populations” (Pernthaler & Amann) somehow subjective, but may reflect better ecologically relevant populations Rosselló-Móra & López-López, 2008. In: Accessing Uncultivated Microorganisms ASM Press López-López et al., 2010 Environ Microbiol Reports 2:258-271 Pernthaler & Amann. 2005. Microbiol Mol Biol Rev 69:440-461 OPUs reduce diversity but may reflect metabolic groups OPUs Reduce diversity measures somehow more tedious and subjective (not always negative) avoid the use of artificial thresholds Species OTUs OPUs Species are taxonomic units based on well characterized isolates Molecular microbial ecology sequences 97% identity = OTU (artificial threshold) I recommend you to use a cutoff value of 99% for OTU clustering OPUs avoid rigid thresholds & may reflect better metabolic and/or ecological types