rcm7504-sup-0001-Supplementary
advertisement

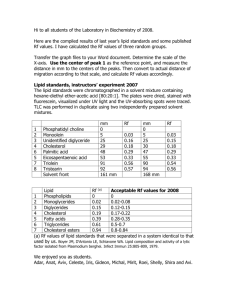
SUPPORTING INFORMATION Culture of Bacillus circulans 344-35 Glucose, magnesium sulfate heptahydrate (MgSO4·7H2O), hydroxy-propyl--cyclodextrin (HPβCD), iron (III) chloride (FeCl3), disodium molybdate (VI) dihydrate (Na2MoO4·2H2O), and copper (II) chloride (CuCl2) were purchased from Nacalai Tesque, Inc. (Kyoto, Japan). Ethyl acetate (EtOAc), sodium sulfate (Na2SO4), calcium chloride dihydrate (CaCl2·2H2O), cobalt chloride (CoCl2), calcium chloride (CaCl2), and boric acid (H3BO3) were purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan). Zinc chloride (ZnCl2), heart infusion agar, and heart infusion broth were purchased from Sigma–Aldrich (St. Louis, MO, USA). Yeast extract was purchased from BD (Franklin Lakes, NJ, USA). Act-1 medium was prepared using the following compounds: 5 g glucose; 10 g yeast extract; 0.6 g dipotassium hydrogen phosphate; 0.3 g potassium dihydrogen phosphate; 0.1 g MgSO4·7H2O; 0.1 g CaCl2·2H2O; 0.05 g NaCl; 14 g HPβCD; 1 mL trace elements; and 1 L sterile water. Trace elements included FeCl3 (27 g), ZnCl2 (1.31 g), CoCl2 (2 g), 1 Na2MoO4 (2 g), CaCl2 (1 g), CuCl2 (1 g), and H3BO3 (0.5 g) in 1 L of sterile water. Bacillus circulans 344-35 was suspended in 100 µL of sterile water and a drop of the solution was incubated overnight in a 10-cm dish containing 8 mL of heart infusion agar at 28°C. After incubation, a single colony was collected and incubated in 10 mL of heart infusion broth at 28°C for 36 h. After incubation, 850 µL aliquots of culture media were preserved with 150 µL of glycerol at −80°C until use for lipid biosynthesis. One milliliter of frozen sample was incubated in 100 mL of Act-1 medium at 28°C for 24 h. After incubation, 1 mL of culture medium was transferred to 20 mL of Act-1 medium, 7.5 µL of 100 µg/µL docosahexaenoic acid (DHA) was added, and bacteria were cultured for 72 h. At 0, 0.5, 1, 2, 3, 24, 48, and 72 h after DHA addition, 100-µL aliquots of culture media were collected and preserved with 400 µL of MeOH at −80°C until lipid extraction. Using 100-µL aliquots of preserved samples, lipid extraction was carried out according to the protocol for “Lipid extraction from mouse lung homogenates” described in the Experimental section of the main article. Reconstituted samples were measured using LC-MS/MS as described in the “Lipid analysis by the API5000 system” section below. Lipid analysis by the API5000 system 2 Lipid analysis was conducted according to our previous report.[1] Briefly, separation was undertaken on an Acquity UPLC system (Waters Corp.) and achieved with an Acquity UPLC BEH C18 column (100 mm × 2.1 mm i.d., 1.7 µm). Mobile phase A was water/1 M ammonium acetate/5 mM phosphoric acid/formic acid (990/10/1/1, v/v/v/v) and mobile phase B was acetonitrile/isopropanol/1 M ammonium acetate/formic acid (495/495/10/1, v/v/v/v). The flow rate was 0.4 mL/min. The gradient program was: 0.00–8.00 min (from 35% to 80% B), 8.01–11.00 min (100% B), and 11.01–13.00 min (35% B). MS analyses were carried out on an API5000 Mass Spectrometer (AB SCIEX, Foster City, CA, USA) equipped with an ESI source. Nitrogen was used as the collision gas for all metabolites. Oxidized fatty acids were detected in negative ESI mode with the following source parameters: curtain gas, 15 psi; ion source gas 1, 50 psi; ion source gas 2, 60 psi; ionspray voltage, −4500 V; collision gas, 8 psi; temperature, 500°C; interface heater, “on”; entrance potential, −10 V; and collision cell exit potential −10 V. Dwell times were set to 25 ms for all molecules to obtain more than 10 data points per peak. Acquisition and processing of data were carried out with Analyst v1.4.2 (AB Sciex). 3 Bioconversion of 21-HDoHE and 22-HDoHE For synthesis of 21-HDoHE, B. circulans 344-35 was incubated with 300 mg of DHA in 3 L of Act-1 medium at 28°C for 7 h. Lipids were extracted with EtOAc (3 L) and evaporated to 20 mL. The organic layers were washed with water, dried over Na2SO4, and concentrated in vacuo. The residue was chromatographed on a Hi-Flash™ ODS column (180 mm × 60 mm i.d.; Yamazen Science Inc., Osaka, Japan) using a linear solvent gradient from 30% acetonitrile:70% water to 60% acetonitrile:40% water over 10 min at 20 mL/min. The synthesis of 22-HDoHE followed a similar protocol to the synthesis of 21-HDoHE. After purification, 21-HDoHE and 22-HDoHE were re-refined using HPLC-UV as described in the “Purification and fractionation of 21-HDoHE and 22-HDoHE using HPLC-UV” section below. Purification and fractionation of 21-HDoHE and 22-HDoHE using HPLC-UV The purification and fractionation of 21-HDoHE and 22-HDoHE were carried out using a HPLC-UV system composed of an LC-8A pump, a SCL-10A system controller and a 4 SPD-10A UV/Vis Detector (Shimadzu Corporation, Kyoto, Japan). The separation of 21-HDoHE was achieved with a Develosil ODS-HG column (150 mm × 20 mm i.d., 5 µm; Nomura Chemical Co., Ltd., Aichi, Japan) and a COSMOSIL -NAP (150 mm × 20 mm i.d., 5 µm; Nacalai Tesque, Inc.) for repurification. The separation of 22-HDoHE was achieved with a Unison US-C18 column (150 mm × 20 mm i.d., 5 µm; Imtakt Corp.). Samples were injected manually. Mobile phases A and B were water and acetonitrile, respectively. The flow rate was 15.0 mL/min. The gradient program was: 0–10 min (50% B) and 10–30 min (from 50% to 70% B). Molecules were detected at a wavelength of 210 nm and the amounts of these molecules recovered were calculated from a calibration curve for DHA. Structure determination of 21-HDoHE and 22-HDoHE using NMR Fractionated 21-HDoHE and 22-HDoHE were subjected to structural analyses by NMR. 1 H and two-dimensional 1H–1H (correlation spectroscopy) NMR spectra of 21-HDoHE and 22-HDoHE in CD3CN were recorded at 302 K on Varian Unity Mercury 300 instruments (300 MHz for 1H NMR). Chemical shifts are denoted in δ (ppm) relative to residual solvent 5 peaks as the internal standard (CD3CN, H1 δ 1.93). 21-HDoHE 1 H NMR (CD3CN): 5.39–5.29 (m, 12H, =C-H), 4.57 (m, 1H, =CH-CH(OH)-CH3), 3.49 (t, 1H, J = 6.8 Hz), 2.87–2.85(m, 10H, =CH-CH2-CH= ), 2.31–2.28 (m, 4H, CH2-CH2-COOH, CH2-CH2-COOH), 1.14 (d, 2H, J = 6.38 Hz). 22-HDoHE 1 H NMR (CD3CN): 5.45–5.31 (m, 12H, =C-H), 3.49 (t, 1H, J = 6.8 Hz), 2.87–2.83 (m, 10H, =CH-CH2-CH=), 2.31–2.22 (m, 6H, CH2-CH2-COOH, CH2-CH2-COOH, =CH-CH2-CH2OH). Evaluation the bioactivity of oxidized fatty acids using NHBECs Normal human bronchial epithelial cells (NHBECs) and bronchial epithelial basal medium (BEBM) were purchased from Lonza Group Ltd. (Basel, Switzerland). NHBECs (2.0×103 cells/well) were incubated in 96-well plates with and without 50 types of oxidized 6 fatty acids at 2 µM or 10 µM concentrations (n = 3) in BEBM at 37°C in a humidified incubator under 5% CO2 for 48 h. A BrdU uptake assay and 3-(4,5-di-methylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay were carried out using a Cell Proliferation Biotrak ELISA system, version 2 (GE Healthcare, Tokyo, Japan) and CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega KK, Osaka, Japan), according to the manufacturers’ instructions. Briefly, for the BrdU uptake assay, BrdU was added to each well (final concentration 10 µM/well) 46 h after the addition of oxidized fatty acids, and plates were incubated for 2 h at 37°C in the humidified incubator under 5% CO2. Subsequently, fixation and blocking were carried out and peroxidase-labeled anti-BrdU was added to each well. After washing, 3,3′,5,5′-tetramethylbenzidine substrate was added to each well and the optical density measured at 450 nm using an EnVision® Xcite Multilabel Reader (Perkin Elmer, Waltham, MA, USA). For the MTT assay, CellTiter 96 AQueous One Solution Reagent was added to each well 48 h after the addition of oxidized fatty acids, and plates were incubated from 1– 2 h at 37°C in the humidified incubator under 5% CO2. After incubation, the absorbance was measured four times at 30-min intervals at 450 nm using the EnVision Xcite Multilabel Reader. The mean values of the measurements were calculated and summarized in a heat 7 map. Glycerolipid, glycerophospholipid, and sphingolipid extraction from lung homogenates Glycerolipid, glycerophospholipid, and sphingolipid extraction was conducted according to a previous report.[2] Briefly, aliquots of lung homogenate solution (10 µL) were transferred to silicone-coated tubes followed by additions of 10 ng/µL IS working solution (10 µL) and water (790 µL). IS working solution included: 16:0-d31 ceramide, d5-(18:0/0:0/18:0) diacylglycerol, 16:0-d31 lysophosphatidylcholine, 13:0 lysophosphatidylinositol, 16:0-d31-18:1 phosphatidic acid, 16:0-d31-18:1 phosphatidylcholine, 16:0-d31-18:1 phosphatidylethanolamine, 16:0-d31-18:1 phosphatidylglycerol, 16:0-d31-18:1 phosphatidylinositol, 16:0-d31-18:1 phosphatidylserine, 16:0-d31 sphingomyelin, sphingosine-d7, sphinanine-d7, sphingosine-1-phosphate-d7, and d5-(17:0/17:1(10Z)/17:0) triacylglycerol. An 800-µL aliquot of 1-butanol was added and the mixture was vortexed vigorously for 10 min at room temperature. After centrifugation at 9,000 × g for 5 min at room temperature, the upper 8 layer was transferred to another silicone-coated tube. The remaining aqueous layer was extracted again with 1-butanol (500 µL). The combined upper layers from both extractions were dried under a gentle stream of nitrogen at 40°C, reconstituted in 200 µL of methanol/chloroform (1/1, v/v), and 1 µL of the reconstituted solution was introduced into the LC-MS/MS system. Glycerolipid, glycerophospholipid, and sphingolipid analysis by Q Exactive Plus system Glycerolipids, glycerophospholipids, and sphingolipids were analyzed as described in the “Lipid analysis by Q Exactive Plus system” in the Experimental section of the main article. The gradient program for HPLC methods was: 0.00–43.00 min (from 40% to 100% B), 43.01–48.00 min (100% B), and 48.01–50.00 min (40% B). Lipid analysis was performed using MS full scan mode ranging from 200 to 1300 in positive ion mode or negative ion mode. The source parameters in positive ion mode were: MS1 resolving power, 70,000; MS2 resolving power, 17,500; capillary temperature, 350°C; auxiliary gas heater temperature, 425°C; ion spray voltage, 3.8 kV; Maximum injection time (IT) of MS1, 100 9 ms; AGC target of MS1, 1×106; Maximum IT of MS2, 50 ms; AGC target of MS2, 5×104; stepped normalized collision energy, 25, 35, and 45 eV; isolation window, 2.0 m/z; apex trigger, between 2 s and 7 s; and dynamic exclusion, 5 s. The X, Y, and Z positions of the heated-ESI probe were set to 0, 2, and C, respectively. Nitrogen was used as the collision gas for all metabolites. The source parameters in negative ion mode followed the analytical method for oxidized fatty acids except for the stepped normalized collision energy; this parameter was set to 25, 35, and 45 eV. The inclusion list, which contains 2053 m/z values of lipid molecular species in the LIPID MAPS database for the positive ion mode, and 2138 m/z values for the negative ion mode, was set to conduct effective fragmentation. The Orbitrap analyzer (Thermo Fisher, Rockford, IL, USA) was calibrated in positive ion mode and negative ion mode according to the manufacturer’s instructions. Identification by Lipid Search Identification of lipid molecular species was performed using Lipid Search v4.1 software (Mitsui Knowledge Industry, Tokyo, Japan). The product search mode was used, and identification was based on the accurate mass of precursor ions and MS2 special 10 patterns. The precursor tolerance was set to a 6.5 ppm mass window and the product tolerance was set to an 8 ppm mass window. The absolute intensity threshold of precursor ions was set to 50,000, and the relative intensity threshold of product ions was set to 1.0%. The m-score threshold was set to 1.0. 11 REFERENCES [1] T. Sanaki, T. Fujihara, R. Iwamoto, T. Yoshioka, K. Higashino, T. Nakano, Y. Numata. Improvements in the High-Performance Liquid Chromatography and Extraction Conditions for the Analysis of Oxidized Fatty Acids Using a Mixed-Mode Spin Column. Mod. Chem. Appl. 2015, 3, DOI:10.4172/2329-6798.1000161. [2] H. Ogiso, R. Taguchi. Reversed-phase LC/MS method for polyphosphoinositide analyses: changes in molecular species levels during epidermal growth factor activation in A431 cells. Anal. Chem. 2008, 80, 9226. 12 Supporting Figure Legends Fig. S1. HPLC/UV and NMR analysis of synthesized 21-HDoHE and 22-HDoHE. HPLC chromatograms of synthesized 21-HDoHE (a) and 22-HDoHE (c), and 1H NMR spectra of synthesized 21-HDoHE (b) and 22-HDoHE (d). Fig. S2. Evaluation of bioactivity of oxidized fatty acids using BrdU uptake and MTT assays. Using NHBECs (2.0×103 cells/well), the cell proliferation activity and cytotoxicity of 50 types of oxidized fatty acids were evaluated at 2 µM or 10 µM (n = 3) by BrdU uptake assay and MTT assay, respectively. The mean values of the measurements were calculated and summarized in a heat map. Fig. S3. Lipid profiling of Bacillus circulans 344-35 after docosahexaenoic acid (DHA) addition. The vertical and horizontal axes are concentration (pg/µL) and time (h), respectively. Samples were collected at 0, 0.5, 1, 2, 3, 24, 48, and 72 h after DHA addition. Collected samples were extracted and analyzed using the API5000 system. The B. circulans 344-35 (DHA+) group, Act-1 medium (DHA+) group, B. circulans 344-35 (DHA−) group, and Act-1 medium (DHA−) group are indicated by purple, red, green, and blue lines, 13 respectively. The mean values and the standard deviations of the measurements (n = 3) for each group were calculated for all lipids. COX: cyclooxygenase, LOX: lipoxygenase, P450: cytochrome P450. 14 Table S1. Number of lipids identified from mouse lung tissue Lipid class Positive ion mode Negative ion mode Ceramide 37 1 Diacylglycerol 146 0 Lysophosphatidic acid 0 4 Lysophosphatidylcholine 150 14 Lysophosphatidylethanolamine 76 25 Lysophosphatidylglycerol 3 14 Lysophosphatidylinositol 2 9 Lysophosphatidylserine 14 18 Phosphatidic acid 8 78 Platelet-activating factor 0 49 Phosphatidylcholine 626 50 Phosphatidylethanolamine 647 230 Phosphatidylglycerol 78 81 Phosphatidylinositol 19 52 Phosphatidylserine 176 190 Sphingomyelin 100 64 Sphingosine 6 0 Triacylglycerol 369 0 Dimethylphosphatidylethanolamine 10 191 Total 2467 1070 15 Figure S1. 16 Figure S2. 17 Figure S3. 18