Lecture 15a
advertisement

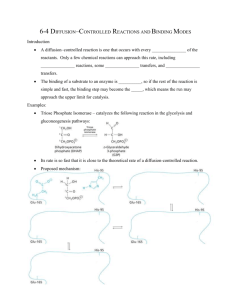
Enzyme Catalysis 3/17/2003 General Properties of Enzymes •Increased reaction rates sometimes 106 to 1012 increase Enzymes do not change DG just the reaction rates. •Milder reaction conditions •Great reaction specificity •Can be regulated Substrate specificity The non-covalent bonds and forces are maximized to bind substrates with considerable specificity •Van der Waals forces •electrostatic bonds (ionic interactions) •Hydrogen bonding •Hydrophobic interaction A+B Substrates enz P+ Q Products Enzymes are Stereospecific O CH3CH 2OH NAD CH3CH NADH H Yeast Alcohol dehydrogenase H O C NH2 NAD+ CH3CD2OH + + N Ox. D H O CH3C-D O C + N NADD Pro-R hydrogen gets pulled off Yeast Alcohol dehydrogenase NH2 Red. OH O 2. NADD + CH3-C-H H C D CH3 O OH 3. CH3-C-D + NADH D C H CH3 If the other enantiomer is used, the D is not transferred YADH is stereospecific for Pro-R abstraction Both the Re and Si faced transfers yield identical products. However, most reactions that have an Keq for reduction >10-12 use the pro-R hydrogen while those reactions with a Keq <10-10 use the pro-S hydrogen. The reasons for this are still unclear Specific residues help maintain stereospecificity Liver alcohol dehydrogenase makes a mistake 1 in 7 billion turnovers. Mutating Leu 182 to Ala increases the mistake rate to 1 in 850,000. This is a 8000 fold increase in the mistake rate, This suggests that the stereospecificity is helped by amino acid side chains. Geometric specificity Selective about identities of chemical groups but Enzymes are generally not molecule specific There is a small range of related compounds that will undergo binding or catalysis. Similar shaped molecules can be highly toxic H O C NH2 N + N N CH3 Tobacco Nicotine Because of this closeness in name Nicotinic acid was renamed to niacin by the bread manufactures Coenzymes Coenzymes: smaller molecules that aid in enzyme chemistry. Enzymes can: a. Carry out acid-base reactions b. Transient covalent bonds c. Charge-charge interactions Enzymes can not do: d. Oxidation -Reduction reactions e. Carbon group transfers Prosthetic group - permanently associated with an enzyme or transiently associated. Holoenzyme: catalytically active enzyme with cofactor. Apoenzyme: Enzyme without its cofactor Commom Coenzymes Coenzyme Reaction mediated Biotin Carboxylation Cobalamin (B12) Alkylation transfers Coenzyme A Acyl transfers Flavin Oxidation-Reduction Lipoic acid Acyl transfers Nicotinamide Oxidation-Reduction Pyridoxal Phosphate Amino group transfers Tetrahydrofolate One-carbon group transfers Thiamine pyrophosphate Aldehyde transfer Vitamins are Coenzyme precursors Vitamin Coenzyme Deficiency Disease Biotin Biocytin Cobalamin (B12) Cobalamin Pernicious anemia Folic acid tetrahydrofolate Neural tube defects Megaloblastic anemia Nicotinamide Nicotinamide Pellagra Pantothenate Coenzyme A Not observed Pyridoxine (B6) Pyridoxal phosphate Not observed Riboflavin (B2) Flavin Not observed Thiamine (B1) Thiamine pyrophosphate Beriberi not observed These are water soluble vitamins. The Fat soluble vitamins are vitamins A and D. Humans can not synthesize these and relay on their presence in our diets. Those who have an unbalanced diet may not be receiving a sufficient supply. Niacin (niacinamide) deficiency leads to pellagra characterized by diarrhea, dermatitis and dementia. Pellagra was endemic is Southern United States in the early 20th century. Niacin can be synthesized from the essential amino acid, tryptophan. A corn diet prevalent at the time restricted the absorption of tryptophan causing a deficiency. Treatment of corn with base could release the tryptophan (Mexican Indians treated corn with Ca(OH)2 before making tortillas!) Regulation of Enzymatic Activity There are two general ways to control enzymatic activity. 1. Control the amount or availability of the enzyme. 2. Control or regulate the enzymes catalytic activity. Each topic can be subdivided into many different categories. Enzyme amounts in a cell depend upon the rate in which it is synthesized and the rate it is degraded. Synthesis rates can be transcriptionally or translationally controlled. Degradation rates of proteins are also controlled. However, We will be focusing on the regulation of enzymatic activity. The catalytic activity of an enzyme can be altered either positively (increasing activity) or negatively (decreasing activity) through conformational alterations or structural (covalent) modifications. Examples already encountered is oxygen, carbon dioxide, or BPG binding to hemoglobin. Also, substrate binding to the enzyme may also be modified by small molecule effectors changing its catalytic site. Protein phosphorylation of Ser residues can activate or deactivate enzymes. These are generally hormonally controlled to ensure a concerted effect on all tissues and cells. Aspartate Transcarbamoylase: the first step in pyrimidine biosynthesis. O O - NH2 O CH2 OPO3-- ATCase + H3N+ Carbamoyl phosphate O C C C O- C H CH2 NH2 H PO 2 4 + C COO Aspartate - O N H C H COO- N-Carbamoyl aspartate This enzyme is controlled by Allosteric regulation and Feedback inhibition Notice the S shaped curve (pink) cooperative binding of aspartate Positively homotropic cooperative binding Hetertropically inhibited by CTP Hetertropically activated by ATP Feedback inhibition Where the product of a metabolic pathway inhibits is own synthesis at the beginning or first committed step in the pathway CTP is the product of this pathway and it is also a precursor for the synthesis of DNA and RNA (nucleic acids). The rapid synthesis of DNA and/or RNA depletes the CTP pool in the cell, causing CTP to be released from ATCase and increasing its activity. When the activity of ATCase is greater than the need for CTP, CTP concentrations rise rapidly and rebinds to the enzyme to inhibit the activity. ATP activates ATCase. Purines and Pyrimidines are needed in equal amounts. When ATP concentrations are greater than CTP, ATP binds to ATCase activating the enzyme until the levels of ATP and CTP are about the same. Enzymatic catalysis and mechanisms •A. Acid - Base catalysis •B. Covalent catalysis •C. Metal ion aided catalysis •D. Electrostatic interactions •E. Orientation and Proximity effects •F. Transition state binding General Acid Base Rate increase by partial proton abstraction by a Bronsted base or Rate increase by partial proton donation by a Bronsted Acid Mutarotation of glucose by acid and base catalysts d D glucose v k obs D glucose dt The reaction can be followed by observation of the optical activity change Kobs = apparent first order kinetics but increases with increased concentrations of acid and base. The acid HA donates a proton to ring oxygen, while the base abstracts a proton from the OH on carbon 1. To form the linear form. The cycle reverses itself after attacking the carbonyl from the other side. This compound does not undergo mutarotation in aprotic solvents. Aprotic solvents have no acid or base groups i.e. Dimethyl sulfoxide or dimethyl formamide. Yet the reaction is catalyzed by phenol, a weak acid and pyridine, a weak base v = k[phenol][pyridine][TM--D-glucose] The reaction can be catalyzed by the addition of -Pyridone as follows v=k'[-pyridone][TM-a-D-glucose] k' = 7000M x k or 1M -pyridone equals [phenol]=70M and [pyridine]=100M Many biochemical reactions require acid base catalysis •Hydrolysis of peptides •Reactions with Phosphate groups •Tautomerizations •Additions to carboxyl groups Asp, Glu, His, Cys, Tyr, and Lys have pK’s near physiological pH and can assist in general acid-base catalysis. Enzymes arrange several catalytic groups about the substrate to make a concerted catalysis a common mechanism. RNase uses a acid base mechanism Two histidine residues catalyze the reaction. Residue His 12 is deprotonated and acts as a general base by abstracting a proton from the 2' OH. His 119 is protonated and acts as a general acid catalysis by donating a proton to the phosphate group. The second step of the catalysis His 12 reprotonates the 2'OH and His 119 reacts with water to abstract a proton and the resulting OH- is added to the phosphate. This mechanism results in the hydrolysis of the RNA phosphate linkage. Covalent catalysis Covalent catalysis involves the formation of a transient covalent bond between the catalyst and the substrate Catalysis has both an nucleophilic and an electrophilic stage 1 Nucleophilic reaction forms the covalent bond 2 Withdrawal of electrons by the now electrophilic catalyst 3 Elimination of the catalyst (almost the reverse of step 1) Depending on the rate limiting step a covalent catalytic reaction can be either elecrophilic or nucleophilic. Decarboxylation by primary amines are electrophilic because the nucleophilic step of Schiff base formation is very fast. Nucleophilicity is related to the basicity but instead of abstracting a proton it attacks and forms a covalent bond. Lysines are common in formation of schiff bases while thiols and imidazoles acids and hydroxyls also have properties that make good covalent catalysts Thiamine pyrophosphate and pyridoxal phosphate also show covalent catalysis Metal ion catalysts One-third of all known enzymes needs metal ions to work!! 1. Metalloenzymes: contain tightly bound metal ions: I.e. Fe++, Fe+++, Cu++, Zn++, Mn++, or Co++. 2. Metal-activated enzymes- loosely bind ions Na+, K+, Mg++, or Ca++. They participate in one of three ways: a. They bind substrates to orient then for catalysis b. Through redox reactions gain or loss of electrons. c. electrostatic stabilization or negative charge shielding Charge stabilization by metal ions Metal ions are effective catalysts because unlike protons the can be present at higher concentrations at neutral pH and have charges greater than 1. Metal ions can ionize water at higher concentrations The charge on a metal ion makes a bound water more acidic than free H2O and is a source of HO- ions even below pH 7.0 NH3 5 Co3 H2O NH3 5 Co3 OH- H The resultant metal bound OH- is a potent nucleophile Carbonic Anhydrase CO 2 H 2O HCO3 H Charge shielding Proximity and orientation effects d p NO2O k1imidizolep NO2Ac k1 p NO2Ac dt k'1 = 0.0018s-1 when [imidazole] = 1M When the phenyl acetate form is used k2 = 0.043 or 24k'1 Proximity effects lead to relatively small rate enhancement! •Reactants are about the same size as water molecules (approximation) •Each species has 12 nearest neighbors (packed spheres) •Reactions only occur between molecules in contact •Reactant conc. Is low so only one can be in contact at a time k1 A B A dA B B v k1AB k 2 A, Bpair dt 12AB A, Bpairs 55.5M 55.5 v k1 A, Bpairs 4.6k1A, Bpairs 12 Only a 4.6 rate enhancement but molecular motions if slowed down leads to a decrease in entropy and rate enhancements. Molecules are not as reactive in all directions and many require proper orientation to react. Increases in rates of 100 fold can be achieved by holding the molecules in their proper orientation for reaction. Preferential transition state binding Binding to the transition state with greater affinity to either the product or reactants. RACK MECHANISM Strain promotes faster rates The strained reaction more closely resembles the transition state and interactions that preferentially bind to the transition state will have faster rates kN S P kE ES EP kN for uncatalyzed reaction and kE for catalyzed reaction K N‡ ‡ E S S E P E KR ‡ KE KT ‡ ES ES ES KR ES ES KE ES ‡ ‡ EP E S ‡ K N‡ ES‡ KT E S ‡ ES K T SES K E ‡ ‡ K R S ES K N ‡ ‡ k BT ‡ k BT ‡ v N k N S S K N S h h k BT ‡ k BT ‡ vE ES K E ES h h ‡ k E K E KT ‡ k N K N KR Preferential transition state binding The more tightly an enzyme binds its reaction’s transition state (KT) relative to the substrate (KR) , the greater the rate of the catalyzed reaction (kE) relative to the uncatalyzed reaction (kN) Catalysis results from the preferred binding and therefore the stabilization of the transition state (S ‡) relative to that of the substrate (S). DGN DGE kE RT exp kN The enzyme binding of a transition state (ES‡ ) by two hydrogen bonds that cannot form in the Michaelis Complex (ES) should result in a rate enhancement of 106 based on this effect alone 106 rate enhancement requires a 106 higher affinity which is 34.2 kJ/mol Transition state analogues are competitive inhibitors