protein synthesis notes

What do all of these have in common?
Collagen
Melanin
Hemoglobin
Lactase
Immunoglobulins
Actin & Myosin
They are ALL types of Proteins that do “work” that contribute to our genetic traits
Protein Synthesis
DNA RNA Proteins
Honors Biology
Ms. Pagodin
Review…
Summarize the structure and function of genes
Describe the function of ribosomes
Differentiate between DNA and genes
Describe the structure and function of DNA
State the base pairing rules
Compare RNA & DNA
RNA (Ribonucleic Acid)
Single Strand of Nucleotides
5 C sugar is ribose
Uses the N base uracil (U) instead of thymine (T)
3 Types:
Messenger RNA (mRNA)
An RNA copy of the gene
Carries and delivers genetic info from nucleus to ribosome
Ribosomal RNA (rRNA)
Components of a ribosome
Site of translation
Transfer RNA (tRNA)
Acts as an interpreter
Translates mRNA into amino acid sequences
All 3 types of RNA are essential for processing information from DNA to proteins.. Gene Expression or Protein
Synthesis
Gene Expression
Organisms traits are determined by proteins
Proteins are assembled according to genes on DNA
DNA can not leave the nucleus, but proteins are made in ribosomes, therefore need an intermediate messenger… RNA
2 stages:
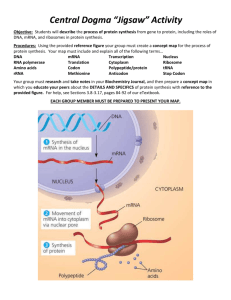
Transcription – copying DNA info to mRNA (nucleus)
Translation – mRNA used to build protein (cytoplasm)
Overview
Transcription
1.
RNA polymerase binds to promoter region of DNA
Promoter region – specific sequence of DNA that serves as a START signal
2.
DNA unwinds and 2 strands separate
only 1 side is used as a template
3.
RNA polymerase reads each nucleotide on the 3’ end and pairs it with a complimentary RNA nucleotide
Same base pairing rules except “U” pairs with “A”
RNA dangles off the enzyme like a tail
4.
Proceeds at 60 nt/sec until RNA polymerase reaches a specific STOP sequence
5.
RNA is released as a free transcript
mRNA Processing
Introns are cut out before mRNA leaves the nucleus
mRNA is a copy of exons (coding) and introns (non-coding) regions
Alternative splicing -
Introns allow for evolutionary flexibility, genes to shuffle, and limits effects of mutations
Add a 5’ cap
Binds to ribosome
Add a 3’ Poly-A tail
100-300 adenine ribonucleotides
Determines how long mRNA will last in the cytoplasm
Compare Transcription to DNA
Replication
Only part of the DNA strand is unwound and used as a template
The enzyme RNA polymerase adds ribonucleotides
Results in a single RNA strand
The Genetic Code
Instructions for building a protein are written as codons on mRNA
Codons – 3 nt that code for a specific a.a.
Codon chart - a.a. and stop signals that are coded by each of 64 possible sequences of mRNA codons
Highly Conserved (Universal) – the genetic code is the same in ALL organisms…significance?
Ex. GUC codes for the a.a. valine in bacteria, dogs, lizards, humans, etc
Reading the codon chart
Translation
tRNA – one loop has 3 nt sequence called an anticodon
Anticodon – 3nt complimentary to codon on mRNA
Enables tRNA to temporarily H-bond to mRNA
No tRNA w/anticodons for STOP codons UAG, UAA, UGA
tRNA also carries the a.a. that corresponds to CODON
Ribosomes
1,00’s in cytoplasm
2 rRNA subunits (large and small) bind together to form ribosome
3 Binding Sites
A site – where tRNA anticodon binds to complimentary codon of mRNA
P site – holds tRNA w/ growing polypeptide chain
E site – tRNA exits, leaving a.a. in the “P” site
Translation: Initiation
Initiator tRNA w/ anticodon
UAC binds to small ribosomal subunit
mRNA start codon binds to tRNA anticodon and finally a large ribosomal subunit binds to the initiation complex
Translation: Elongation
Translation: Termination
Translation: Assembling the
Protein
1.
2.
mRNA binds to small rRNA subunit w/start codon,
AUG, in the “P” site tRNA w/ anticodon UAC and carrying a.a. methionine binds to start codon
3.
The next codon, in “A” site, binds w/ complimentary tRNA (carrying the corresponding a.a.)
4.
Enzyme forms a peptide bond between adjacent a.a.
5.
tRNA in “P” site now exits via “E” site and is recycled
6.
tRNA in the “A” site moves to the “P” site w/ growing polypeptide chain, mRNA moves w/it, therefore a new codon is in the “A" site
7.
Process continues until it reaches a STOP codon at the end of the mRNA, there is no anticodon
8.
W/nothing in the “A” site, the ribosome is disassembled and the newly made polypeptide is released
Protein Synthesis
Mutations
Mutation – any change in an organism’s genetic material
Causes
Mutagens – environmental agents that cause mutations after exposure
X-rays, UV rays, chemicals
Carcinogens – mutagens that lead to cancer
Asbestos, benzene, tobacco
Types of Mutations
Chromosomal Mutations
Alterations in chromosome structure
Deletion, duplication, inversion, translocation
Point Mutations
Just one or a few nt changed in a gene
Substitution – one nt is replaced by a different nt
Ex. UGU UGC (no effect b/c both code for cysteine)
UGU UGA (early STOP codon)
Frameshift mutations
Mutations that cause a gene to be read in the wrong 3 nt sequence
Insertions – one or m ore nt added to gene
Ex. AAU CGC UUU
AGA UCG CUU U
Deletions – one or more nt deleted from gene
Ex. AAU CGC UUU
AUC GCU UU
Note
* If mutation occurs in an intron it will have no effect
*if reading frame is displaced 3 nt, the mutation may have no effect
Prokaryotic Gene Regulation
Prokaryotic Cells – genes are unbroken set of nt
Operon
controls gene expression in prokaryotes
Cluster of genes that code for proteins w/related functions
Lac Operon
Lac Operon – genes for lactose digesting enzyme
Only want lactose digesting enzymes when lactose is present…or else energy is being wasted transcribing genes
Operator – acts like an on/off switch
If no molecule is bound to operator, then the gene is “ON” and RNA polymerase can move across
When a repressor protein binds to the operator, it blocks the RNA polymerase from transcribing, genes are “OFF”
Repressor can be removed by inducer (ex. allolactose), now gene is turned ‘ON”
Trp Operon – genes for making tryptophan
E.coli would typically get trp from environment, therefore gene only needs to be turned on when trp is not present
Trp Operon
Eukaryotic Gene Regulation
No operons…b/c genes w/similar functions are scattered among different chromosomes
Multicellular organisms have different types of cells, all somatic cells contain the same DNA…but what makes them different is which genes are turned on/off
Ex. Every cell has hemoglobin genes, but only turned “ON” in rbc
Transcription takes place at uncoiled regions of chromosome
RNA polymerase cannot bind w/o transcription factors
Transcription factors are signaled by 20 messengers that bind to the enhancer site to turn “ON” the gene