Figure S2. Homology modelling of AAT1-RGa.
advertisement

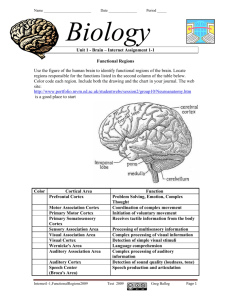
Supporting Information The AAT1 locus is critical for the biosynthesis of esters contributing to ‘ripe apple’ flavour in ‘Royal Gala’ and ‘Granny Smith’ apples Edwige J.F. Souleyre, David Chagné, Xiuyin Chen, Sumathi Tomes, Rebecca M. Turner, Mindy Y. Wang, Ratnasiri Maddumage, Martin B. Hunt, Robert A. Winz, Claudia Wiedow, Cyril Hamiaux, Susan E. Gardiner, Daryl D. Rowan, Ross G. Atkinson Figure S1. Western analysis of AAT1 after transient expression in tobacco leaves and in GS and RG ripe fruit cortex and skin. (A) Proteins (15 g per lane) from Nicotiana benthamiana leaves were extracted after infiltration with vectors transiently expressing Gus (negative control), P19 (negative control), AAT1-RGa or AAT1-GSa. Proteins were separated on 4-12% NuPage gels (Invitrogen) and transferred to PVDF membrane (BioRad). AAT1 proteins were detected using a polyclonal antibody to AAT1-RGa. Blots were incubated with an alkaline phosphatase conjugated antirabbit secondary antibody (Sigma-Aldrich at 1:1000 w/v) and AAT1 binding was visualised using 1-Step NBT/BCIP (Pierce) as described in Maddumage et al., 2013 (J. Agric. Food Chem. 61, 728-739). L = SeeBlue Plus2 Pre-Stained Protein ladder (Invitrogen), sizes in kDa. (B) Proteins from GS and RG ripe fruit skin (sk) and cortex (cx) tissues were extracted using 70 l of hot SDS buffer according to Maddumage et al., 2013. Proteins were separated on 12% TrisTricine gels. AAT1 binding was detected as described above. The AAT1 antibody was raised to recombinant AAT1-RGa protein produced as follows. AAT1-RGa was subcloned into the E. coli expression vector pET32Xa/LIC using the pET Ligation Independent Cloning System (Novagen) resulting in the clone pET32Xa/LIC-AAT1-RGa (Souleyre et al., 2005). Recombinant N-terminal His-tagged protein was expressed and purified by Ni2+ affinity chromatography under denaturing conditions according to manufacturer’s instructions (GE Healthcare). Purified insoluble recombinant AAT1 protein (~500 µg) was used to raise a polyclonal antibody in rabbit (AgResearch, NZ). Figure S2. Homology modelling of AAT1-RGa. Two closely related hydroxycinnamoyltransferases (HCT, from sorghum and coffee) were identified as the most suitable templates for AAT1-RGa by the HHPred server (Söding et al., 2005, Nucleic Acids Research 33, 244-248). Like AAT1-RGa, these two proteins belong to the clade V of the BADH superfamily (D’Auria, 2006) and their sequence identity with AAT1-RGa is 28%. The crystal structure of the sorghum HCT in complex with reaction products p-coumaroyl shikimate and HS-CoA (PDB entry 4KEC) was used as template for homology modelling of AAT1-RGa. Sequences were aligned using the Clustal W2 server (Larkin et al., 2007, Bioinformatics 23, 2947-2948) and modelling was done using Modeller (Eswar et al., 2006, Current Protocols in Bioinformatics 15, 5.6.1-5.6.30). The catalytic His164 in the AAT1-RGa model is located in the middle of the solvent channel running between the two domains. Asp168, from the conserved HXXXD motif, is engaged in the conserved salt bridge with Arg296, a feature proposed to be critical for maintaining proper interdomain distance and conformation (Ma et al., 2005, J. Biol. Chem. 280, 13576-13583; Walker et al., 2013, Plant Phys. 162, 640-651). The three key amino acid differences between RG and GS are indicated within the orange ellipse which shows the approximate location of the alcohol binding site. Figure S3. AAT1-RGa gene-specific expression profiles in transgenic RG apple and control lines. Relative gene expression for AAT1-RGa was determined by qRT-PCR in both skin (S) and cortex tissues (C) of RG control and transgenic lines AS70, AS1996, AS2002 and AS2004. Skin and cortex tissues were sampled at ripe stage (150 days after full bloom, starch index ~6). Gene expression data analysis was carried out as for Figure 3. Data are presented as mean ± SEM (n=4). Figure S4. Expression profiles of genes from fatty acid and isoleucine biosynthesis pathways in transgenic RG apple and control lines. Relative gene expression was determined by qRT-PCR in both skin (S) and cortex tissues (C) of RG control and the transgenic lines AS70 and AS2004. Skin and cortex tissues were sampled at ripe stage (150 days after full bloom, starch index ~6). Gene expression profiles are shown for (A) carboxylesterase MdCXE1, (B) carboxylesterase MdCXE16, (C) lipoxygenase LOX1, (D) lipoxygenase LOX7, (E) threonine synthase TS1 and (F) branched chain aminotransferase BCAT1. Gene expression data analysis was carried out as for Figure 3. qRT-PCR were performed in triplicate using cDNA synthesized from three individual RNA extractions. Data are presented as mean ± SEM (n=12). There is significant differences if treatment shown a different letter. The letters given by statistical analysis only apply when comparing the three treatments with the same sample type (cortex or skin).