BIO 330 Cell Biology Lecture Outline Spring 2011 Chapter 10
advertisement

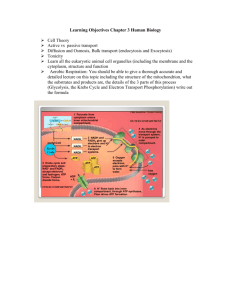
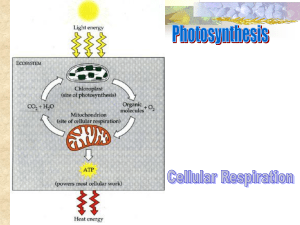
BIO 330 Cell Biology Lecture Outline Spring 2011 Chapter 10: Aerobic Respiration I. Cellular Respiration: Overview A. Aerobic vs anaerobic respiration B. Aerobic respiration vs fermentation C. Overall process Glycolysis Pyruvate oxidation to Acetyl CoA TCA cycle Electron transport ATP synthesis II. The Mitochondrion A. Mitochondrial location B. Structure Two membranes (outer vs inner) Three regions Intermembrane space Matrix Intracristal spaces C. Functional / structural relationship D. Bacteria lack mitochondria Similar processes occur at the plasma membrane III. The Tricarboxylic Acid Cycle A. Oxidation of pyruvate to Acetyl CoA Oxidative decarboxylation Pyruvate dehydrogenases (PDH) Produces 1 CO2 and 1 NADH per pyruvate B. Acetyl CoA enters the TCA cycle Citrate synthase forms citrate Aconitase converts citrate to isocitrate C. Oxidative decarboxylations Isocitrate dehydrogenase removes CO2 and forms oxalosuccinate, then -ketoglutarate -ketoglutarate dehydrogenase removes CO2 and forms succinyl CoA D. ATP production Co A is released; energy is used to generate ATP or GTP E. NADH and FADH2 generation & oxaloacetate regeneration Succinate dehydrogenase forms fumarate and FADH2 Fumarate hydrates forms malate Malate dehydrogenase forms oxaloacetate and NADH F. Summary of TCA cycle BIO 330 Cell Biology Lecture Outline Spring 2011 Acetyl CoA + 3NAD+ + FAD + ADP + Pi 2CO2 + 3NADH + FADH2 + CoA—SH + ATP For each glucose molecule: 6 NADH, 2 FADH2, and 2 ATP are produced For each glucose molecule, including glycolysis, Acetyl CoA formation, and TCA cycle: 10 NADH, 2 FADH2, 4 ATP G. Regulation of TCA cycle Allosteric regulation of TCA cycle enzymes NADH, ATP, Acetyl CoA are allosteric regulators PDH regulation by phosphorylation H. Lipid and protein catabolism; and protein and nucleic acid anabolism via TCA cycle Amphibolic cycle IV. Electron Transport A. Electron transport system overview Electrons are transferred from NADH and FADH2 to O2 Creates water Releases free energy Electron transfer occurs in stepwise fashion to maximize efficiency B. Five kinds of electron carriers are parts of respiratory complexes Flavoproteins Carry electrons and protons together Iron-sulfur proteins Carry only one electron by redox of iron ions Cytochromes Contain heme; carry only one electron Copper-containing cytochromes Fe-Cu center holds O2 until 4 electrons are received Coenzyme Q (a quinone; aka ubiquinone) Accepts both electrons and protons; transfers protons across membrane C. Sequence of electron transport The position of each carrier is determined by its standard reduction potential Electrons enter either via Complex I or Complex II NADH donates to Complex I FADH2 donates to Complex II Complexes I and II both pass electrons to CoQ, which then passes them to Complex III Complex III passes electrons to Cyt c, which then passes them to Complex IV Complex IV passes electrons to O2 as the terminal electron acceptor V. The Electrochemical Proton Gradient A. Chemiosmotic Model Proton gradient across the inner mitochondrial membrane links electron transport with ATP production BIO 330 Cell Biology Lecture Outline Spring 2011 B. Energy yields Each NADH provides enough energy to produce 3 ATP Each FADH2 provides enough energy to produce 2 ATP C. Proton motive force VI. ATP Synthesis A. F0F1 complex F0 is a membrane-bound proton channel subunit (proton translocator) F1 is an ATPase / ATP synthase subunit attached to F0 B. F0 structure a, b, c subunits a is the proton channel b connects F0 to F1 multiple c subunits form a rotating ring that turns the subunit of F1 C. F1 structure helps is catalytic site for ATP synthesis is rotating stalk that turns inside of the 3 3 wheel immobilizes 3 3 wheel anchors subunit to c ring of F0 D. Binding change model of ATP synthesis subunit has 3 different conformations, each with different affinities for ADP and ATP L (loose) – binds ADP and and Pi loosely T (tight) – binds ADP and Pi tightly and catalyzes condensation O (open) – low affinity for both ADP and ATP