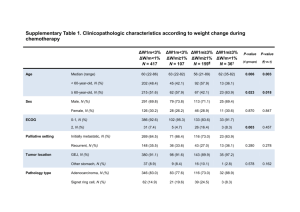
STelesco_HER2
advertisement
Structural dynamics of HER2 and ErbB4: Yin and Yang in Mammary Carcinoma Shannon Telesco Advisor: Ravi Radhakrishnan, Ph.D. Department of Bioengineering The HER2 signaling network Human epidermal growth factor receptor 2 (HER2) is a member of the ErbB family of receptor tyrosine kinases (RTK). Ligand binding induces receptor dimerization and phosphorylation of tyrosine residues in the C-terminal tail segments. Tyrosines serve as docking sites for signaling molecules, activating molecular pathways such as cellular proliferation. Overexpression of HER2 results in ligand-independent activation and occurs in 2030% of human breast cancers. Yarden, Y and Sliwkowski, MX. Untangling the ErbB signalling network. Nat Rev Molecular Cell Biology 2001; 2:127-137. Activation of ErbB tyrosine kinases • ErbB kinases are transmembrane receptors comprised of a ligand-binding extracellular domain, transmembrane segment, intracellular kinase domain, and tyrosine-rich C-terminal tail. • Receptors can be auto- or transphosphorylated in their C-tails. Zhang, X., Gureasko, J., Shen, K., Cole, P., and Kuriyan, J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 2006; 125:1137-1149. Structure of the HER2 kinase domain Alpha C helix: Facilitates coordination of substrate tyrosine Nucleotide-binding loop (N-loop): Coordination of ATP & substrate tyrosine Activation loop (A-loop): Regulates accessibility of active site to binding Catalytic loop: Directly participates in phosphoryl transfer Regulation of HER2 activation αC helix rotates into the active site Y877 Active A-loop αC helix N-loop C-loop Phosphorylation of Y877 in the A-loop may regulate extension of the loop and activation of HER2. Inactive A-loop αC helix N-loop C-loop Elucidating HER2 activation mechanism Investigate the structural differences between inactive and active HER2. What are the key bonds that must be formed or broken upon activation? Define the role of Y877 phosphorylation in HER2 activation. Is HER2 unique from other ErbB members in that P-Y877 is necessary for activity? Predict the behavior of an EGFR/HER2 heterodimer. How might the dimerization interface trigger conformational changes in HER2? Applying molecular dynamics (MD) to the HER2 system Four systems created: HER2 inactive & active, with & without Y877-phosphorylation. Systems were solvated & ionized (150 mM NaCl) & heated to 300 K. MD simulations performed for 10 ns. Trajectories analyzed for key hydrogen bonds and conformational changes. Solvated inactive HER2. Hydrogen bonds in the A-loop Inactive HER2 Y877 Unphosphorylated Salt Bridges Active HER2 Y877 Unphosphorylated D863, K753 Inactive HER2 Y877 Phosphorylated Active HER2 Y877 Phosphorylated D863, K753 E876, R898 D880, R897 D880, R897 Inactive HER2 Y877 Unphosphorylated Active HER2 Y877 Unphosphorylated K883, E766 Inactive HER2 Y877 Phosphorylated Active HER2 Y877 Phosphorylated F864 HN, E770 OE2 G865 HN, V842 O G865 HN, H843 O Conserved bond L866 O, R844 HE/HH11 L866 O, R844 HE/HH11 R868 HH12/22, D769 OD1/2 R868 HH12, R840 O Hydrogen Bonds R868 HN/O, V842 O/HN R868 HN/O, V842 O/HN L870 HN, R840 O L870 HN, R840 O D871 O, R840 HE/HH11/HH12 D873 OD1/2, R897 HE/HH22 E874 OE1/2, T759 HN/HG1 E876 OE1/2, R898 HH22/HE Y877 O2/O3, R844 HH/HH12/HH22 Y877 O2/O3, R844 HH12/HH22 Y877 O3, K883 HZ1/2/3 Y877 O2, K883 HZ1/2/3 Y877 O2, R897 HH12/HH21 Y877 O3, R868 HH21/22 Y877 HN/O, F899 O/HN A879 HN, R897 O K883 HZ1/2/3, E757 OE1/2 A-loop V884 O, K887 HN Hydrogen bonds in the αC helix Inactive HER2 Y877 Unphosphorylated Active HER2 Y877 Unphosphorylated Inactive HER2 Y877 Phosphorylated Active HER2 Y877 Phosphorylated E766, R756 Salt Bridges E766, K883 E770, K753 Inactive HER2 Y877 Unphosphorylated N764 HN, S760 O Key salt bridge Active HER2 Y877 Unphosphorylated E770, K753 Inactive HER2 Y877 Phosphorylated Active HER2 Y877 Phosphorylated A763 HN, S760 OG A763 HN, S760 OG N764 HN, S760 O N764 HN, S760 O E766 OE1/2, R756 HH12/21/22/HE D769 OD1/2, R868 HH12/22 Hydrogen Bonds E770 OE2, F864 HN Y772 O, G776 HN Y772 O, G776 HN V773 O, V777 HN M774 O, L785 HN αC helix V773 O, V777 HN M774 O, L785 HN Dual inhibition of the active state R868 V842 K753 E770 D863 (coordinating Asp) Key salt bridge Inactive HER2: E770-K753 bond is inhibited Active HER2: Sequestering residues release E770 & K753 Stabilizing H-bonds in the active state Active A-loop Inactive A-loop Active αC helix Inactive αC helix K883 E766 • Key salt bridge in active HER2 is K883-E766 • Connects the αC helix with the A-loop, stabilizing the helix in the active site • Bond is conserved among ErbB family members: K851-E734 (EGFR), K856-E739 (ErbB4) Inactive/active HER2 (superimposed) Analysis of conformational shifting RMSD for A-loop and αC helix (10 ns) RMSD (αC helix) 18 16 14 12 10 8 6 4 2 0 12 UnP-Active P-Active UnP-Inactive P-Inactive RMSD WRT Active (Å) RMSD WRT Active (Å) RMSD (A-loop) 10 UnP-Active 8 P-Active 6 UnP-Inactive 4 P-Inactive 2 0 0 2 4 6 8 10 RMSD WRT Inactive (Å) 12 14 0 2 4 6 8 10 RMSD WRT Inactive (Å) • All four systems are stable during 10 ns production run • Slight movement of αC helix in active system •No shifting of inactive toward active in short timescale Effect of Y877 phosphorylation • Many receptor tyrosine kinases, including the insulin receptor, require phosphorylation of their A-loops for full kinase activity. • The EGFR family is unique in that A-loop phosphorylation appears to be unnecessary for activation. • The role of A-loop phosphorylation in HER2 is controversial, as several studies have highlighted the importance of Y877 phosphorylation for kinase activity. Active A-loop Y877 Inactive A-loop Effect of Y877 phosphorylation • Equilibrated P-active HER2 contains a network of hydrogen bonds which pin the A-loop to underlying regions of the kinase, maintaining the loop in its extended conformation. • These fastening residues occur at both ends of the A-loop. R868 L870 R840 Y877 L866 A879 V842 E876 R898 R844 N-terminal end of A-loop R897 F899 C-terminal end of A-loop Role of Y877 in bridging the A-loop R868 • The phosphoryl group on Y877 bridges the fastening residues on either side of the A-loop. • P-Y877 forms hydrogen bonds with residues at the N-terminal end of the A-loop, including K883, R844, and R868. R844 K883 P-active HER2 (Activation loop) • Unphosphorylated active HER2 lacks this bridging mechanism. P-Y877 Comparison between HER2 and insulin receptor tyrosine kinase HER2 (P-active) Insulin RTK P-Y1163 (IRK) P-Y877 (HER2) R1155 (IRK) R868 (HER2) • Equilibrated P-active HER2 shares structural features with P-IRK. • R1155 and P-Y1163 make VDW contacts in IRK. Likewise, R868 and P-Y877 form hydrogen bonds in HER2. • Structure is unique to HER2 & IRK, as R868 is a lysine (K836) in EGFR. P-active HER2 superimposed on insulin RTK Dimerization of HER2 • ErbB kinases dimerize in an asymmetric head-to-tail configuration, similar to that seen for cyclin/cyclindependent kinase complexes. • Monomer A is the activated kinase & monomer B is the activating kinase. • The αC helix of monomer A comprises a key region of the dimerization interface. Monomer B Monomer A Zhang, X., Gureasko, J., Shen, K., Cole, P., and Kuriyan, J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 2006; 125:1137-1149. Constructing an EGFR/HER2 heterodimer • Two different heterodimers were constructed: • Y877-unphosphorylated inactive HER2 (activated kinase), active EGFR (activating kinase) • Y877-phosphorylated inactive HER2 (activated kinase), active EGFR (activating kinase) • Systems were solvated & ionized (150 mM NaCl) & heated to 300 K. MD simulations performed for 10 ns. • Does dimerization promote activation of HER2? Is dimerization sufficient for activation or must HER2 also be phosphorylated? HER2 EGFR The dimerization interface HER2: P707 Q711 M712 I714 L768 L790 V794 V794 EGFR: I917 Y920 M921 V924 M928 I929 V956 αC helix Y920 P707 I917 N-terminal tail HER2 Dimer interface EGFR Conformational shifts in heterodimer RMSD for dimeric HER2: αC helix and A-loop (10 ns) RMSD for Dimer (αC helix) 14 18 16 14 12 10 8 6 4 2 0 P-Y877 Dimer UnP-Y877 Dimer RMSD WRT Active (Å) RMSD WRT Active (Å) RMSD for Dimer (A-loop) 12 10 8 P-Y877 Dimer 6 UnP-Y877 Dimer 4 2 0 0 2 4 6 8 10 12 RMSD WRT Inactive (Å) 14 16 0 2 4 6 8 10 12 14 RMSD WRT Inactive (Å) • No significant movement of HER2 toward active form • Shifting of αC helix due to adjustment to dimeric interface Effect of dimerization on hydrogen bonding network Residues in each of the four monomeric systems predicted to be affected by the dimerization interface: Inactive HER2 Y877 Unphosphorylated Monomer A residues P707 Q711 M712 I714 L768 L790 V794 Active HER2 Y877 Unphosphorylated Y772 Y772, L785 N764, Y772 N764 N764 Inactive HER2 Y877 Phosphorylated Active HER2 Y877 Phosphorylated Y772, G776 Y772, L785 N764, D769 Y772 N764, E770 N764 T759 N764 Bonds broken (Phosphorylated inactive HER2): M774-L785, E874-T759, G865-H843, R868-R840, and V884-K887 Bonds broken (Unphosphorylated inactive HER2): N764-S760, Y772-G776, G865-V842, D873-R897, and K883-E757 Conclusions • Inactive and active HER2 structures reveal distinctive hydrogen bonding patterns that stabilize each conformation. A dual inhibitory mechanism maintains the inactive state through sequestration of key residues required for activation. • Phosphorylation of Y877 may serve to bridge the stabilizing hydrogen bonds on either side of the A-loop in the active conformation. Unphosphorylated active HER2 lacks this bridging mechanism. • Formation of EGFR/HER2 heterodimer results in repositioning of the αC helix and breakage of several key bonds that are present in the inactive state. Part II: Role of ErbB4 signaling in the mammary gland Opposing roles of HER2 and ErbB4 in breast cancer • In contrast to HER2, expression of ErbB4 in breast cancer is associated with a favorable prognosis & a differentiating tumor phenotype. • ErbB4 activation of STAT5a in the mammary gland regulates lactational expression of milk genes such as beta-casein. • STAT5a is recruited to ErbB4 through binding of phosphotyrosine peptides by the SH2 domain. Williams, C., Allison, J.G., Vidal, G.A., Burow, M.E., Beckman, B.S., Marrero, L., and Jones, F.E. The ErbB4/HER4 receptor tyrosine kinase regulates gene expression by functioning as a STAT5a nuclear chaperone. JCB 2004; 167(3): 469-478. PAINTing a picture of the interaction between ErbB4 and STAT5a • Goal is to connect ErbB4 to its regulated transcription factors, such as STAT5a, by applying a bioinformatics program called PAINT. • PAINT (Promoter Analysis & Interaction Network Toolset) is a computational tool which analyzes microarray data & generates networks connecting upregulated genes to their respective transcription factors. • Given a list of genes (microarray data), PAINT can: Fetch potential promoter sequences for the genes in the list. Find Transcription Factor (TF) binding sites on the sequences. Analyze the TF-binding site occurrences for over/under-representation compared to a reference. Vadigepalli, R, Chakravarthula, P, Zak DE, Schwaber JS, and Gonye, GE. PAINT: a promoter analysis and interaction network generation tool for gene regulatory network identification. OMICS 2003; 7(3):235-53. Bridging the genomics and atomistic scales in ErbB4 signaling Input microarray data from experiment Perform PAINT analysis to identify relevant TFs (Genomics Scale) Analyze key TF/binding partner interactions using molecular dynamics (Atomistic Scale) Validate structural predictions experimentally Preliminary results: PAINT analysis • The PAINT method was applied to the following study, which focused on stimulation of ErbB4 in mammary epithelial cells: Amin, DN, Perkins AS, and Stern DF. Gene expression profiling of ErbB receptor and ligand-dependent transcription. Oncogene 2004 Feb 19; 23(7):1428-38. • In the study, agonistic antibodies as well as natural ligands (neuregulin) were used to activate the ErbB4 pathway, and ErbB4-stimulated gene expression was assessed by microarray analysis. • Several novel ErbB4 gene targets were identified and their associated transcription factors were predicted by PAINT. Preliminary results: PAINT analysis Red blocks correspond to over-representation of a TF in a given gene cluster. Cyan blocks correspond to under-representation of a TF in a given gene cluster. TF Genes (Color-Coded by Cluster) STAT5a STAT5a interaction with ErbB4 IFN-γ phosphopeptide • The SH2 domain of STAT5a binds to P-Y959 at the C-terminal end of ErbB4’s kinase domain. • Structural details of the SH2 domain-phosphotyrosine peptide interaction are known (STAT1-IFN-γ crystal structure). • Can we predict features of the interaction between ErbB4 and STAT5a? STAT1 SH2 domain Mao, X., et al. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol Cell 2005; 17(6):761-71. STAT1 bound to phosphotyrosine peptide on IFN-γ receptor. STAT5a interaction with DNA • Upon dimerization, STAT5a migrates to the nucleus and initiates transcription of genes containing GAS promoter sequences. SH2 domains • Crystal structures of STAT1 and STAT3 bound to DNA reveal a nine base-pair consensus sequence. • Monomers form a ‘pliers’-like structure in which dimerization is mediated by the SH2 domains. Chen, X., et al. Crystal structure of a tyrosine phosphorylated STAT1 dimer bound to DNA. Cell 1998; 93(5):827-39. DNA Predicting STAT5a interaction with ErbB4 and DNA • To summarize, goals of ErbB4 study are two-fold: • Predict binding of ErbB4 to STAT5a SH2 domain. Is it structurally possible for ErbB4 to phosphorylate the key tyrosine on STAT5a? Does ErbB4 bind to STAT5a as a monomer or as a dimer? • Assess STAT5a binding to DNA consensus sequence. Which interactions regulate specificity of nucleotide-binding? What mutations in the DNA sequence or STAT5a DNA-binding domain abolish the interaction? • Experimentally validate through mutagenesis studies and EMSA assays. • Elucidate structural features of key interactions involved in the ErbB4 signaling pathway. Acknowledgments Ravi Radhakrishnan, Ph.D. Mark Lemmon, Ph.D. Rajanikanth Vadigepalli, Ph.D. Andrew Shih References Bose, R, Molina, H, Patterson, AS, Bitok, JK, Periaswamy, B, Bader, JS, Pandey, A, and Cole, PA. Phosphoproteomic analysis of Her2/neu signaling and inhibition. PNAS 2006; 103(26):9773-9778. Chen, X, Vinkemeier, U, Zhao, Y, Jeruzalmi, D, Darnell, JE Jr, and Kuriyan, J. Crystal structure of a tyrosine phosphorylated STAT1 dimer bound to DNA. Cell 1998; 93(5):827-39. Fiser, A, Sali, A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol 2003; 374:461-91. Hubbard, SR, Wei, L, Ellis, L, and Hendrickson, W.A. Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature 1994; 372: 746-54. Lemmon, MA. The EGF receptor family as therapeutic targets in breast cancer. Breast Dis 2003; 18:33-43. Linggi, B, Cheng, QC, Rao AR, and Carpenter G. The ErbB4 s80 intracellular domain is a constitutively active tyrosine kinase. Oncogene 2006; 25: 160-63. Liu Y, Purvis J, Shih A, Weinstein J, Agrawal N, Radhakrishnan R. A multiscale computational approach to dissect early events in the Erb family receptor mediated activation, differential signaling, and relevance to oncogenic transformations. Ann Biomed Eng 2007; 35(6): 1012-25. Mao, X, Ren, Z, Parker, GN, Sondermann, H, Pastorello, MA, Wang, W, McMurray, JS, Demeler, B, Darnell, JE Jr, and Chen, X. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol Cell 2005; 17(6):761-71. Schulze, WX, Deng, L, and Mann, M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol Systems Biology 2005; 1:2005.0008. Stamos, J, Sliwkowski, MX, and Eigenbrot, C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. JBC 2002; 277(48): 46265-46272. References Stein, RA and Staros, JV. Insights into the evolution of the ErbB receptor family and their ligands from sequence analysis. BMC Evol Biol 2006; Oct 6; 6:79. Sundvall, M, Peri, L, Maatta, JA, Tvorogov, D, Paatero, I, Savisalo, M, Silvennoinen, O, Yarden, Y and Elenius K. Differential nuclear localization and kinase activity of alternative ErbB4 intracellular domains. Oncogene 2007; 26(48): 6905-14. Vadigepalli, R, Chakravarthula, P, Zak DE, Schwaber JS, and Gonye, GE. PAINT: a promoter analysis and interaction network generation tool for gene regulatory network identification. OMICS 2003; 7(3):235-53. Wang, SE, Narasanna, A, Perez-Torres, M, Xiang, B, Wu, FY, Yang, S, Carpenter, G, Gazdar AF, Muthuswamy, SK, and Arteaga, CL. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell 2006; 10: 25-38. Williams, C., Allison, JG, Vidal, GA, Burow, ME, Beckman, BS, Marrero, L., and Jones, FE. The ErbB4/HER4 receptor tyrosine kinase regulates gene expression by functioning as a STAT5a nuclear chaperone. JCB 2004; 167(3): 469-478. Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, Ellis B, Pennisi C, Horne E, Lackey K, Alligood KJ, Rusnak DW, Gilmer TM, and Shewchuk L. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res 2004; 64(18): 6652-9. Yarden, Y and Sliwkowski, MX. Untangling the ErbB signalling network. Nat Rev Molecular Cell Biology 2001; 2:127-137. Zhang, X., Gureasko, J., Shen, K., Cole, P., and Kuriyan, J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 2006; 125:1137-1149.