Chapter 6 Protein Function Part 2: Enzymes
advertisement

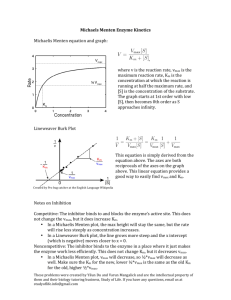
Chapter 6 Protein Function : Enzymes Part 1 Enzymes Enzyme Learning Goals: to Know – Physiological significance of enzymes – Catalytic power of enzymes – Chemical mechanisms of catalysis – Mechanism of chymotrypsin – Description of enzyme kinetics and inhibition Enzymes Mostly Proteins (a few RNA’s are capable of catalysis) Active Site: Substrate Binding + Reaction Products Some require Cofactors (metals) or Coenzymes (organic cpds) Some enzymes have other binding sites…involved in regulation, we will see later, Part 2 EOC Problem 1 involves the sweetness of corn affected by corn enzymes and Problem 2 calculates the average molar concentration of enzymes in a bacterial cell: you can take it further to find the number of molecules of each enzyme present in a cell. Enzyme Pioneers First Cell Free Prep First to Crystallize Urease Weak bonding at active site results in catalysis Why biocatalysis over inorganic catalysts? • • • • Greater reaction specificity: avoids side products Milder reaction conditions: conducive to conditions in cells Higher reaction rates: in a biologically useful timeframe Capacity for regulation: control of biological pathways COO - COO NH2 O OH COO OH COO Chorismate mutase COO OOC O NH2 - - O COO COO • Metabolites have many potential pathways of decomposition - • Enzymes make the desired one most favorable OH EOC Problem 4: Examines the thermal protection of hexokinase that a substrate brings to the table: maintaining conformation under harsh conditions. Later in Part 2 of this chapter we will see X ray data backing this up. Enzymatic Substrate Selectivity: Phenylalanine hydroxylase OH H - H + NH3 OOC - H - + NH3 No binding + NH3 OOC OOC OH HO H H H NH CH3 OH Binding but no reaction Class Is the First Part of E.C. Number EC 2.7.1.1 = ATP:glucose phosphotransferase or Hexokinase 2 = Transferase 7 = Phosphotransferase 1 = Transferred to a hydroxyl 1 = Glucose is the acceptor Enzyme Search By Class Bacterial Luciferase Rxn FMNH2 + O2 + RCHO FMN + RCOOH + H2O + light Continuing with the EC Numbers-1 Continuing with EC Numbers-2 NiceZyme Enzyme with an Active Site Active Site Chymotrypsin Thermodynamics of a Reaction S + E ES E + P Enzyme Catalyzed Reaction E+S ↔ ES ↔ EP ↔ E+P EOC Problem 3: A rate enhancement problem using Urease, the enzyme that converts: Urea CO2 + 2 NH3. The calculation demonstrates how long it would take if urease were not present ! Dihydrofolate Reductase Substrate Binds in a Fold or Pocket NADP+ + Folic Acid ΔGB = binding energy Enzyme Reactions Bind Substrate then Change Shape to Transition State Triose Phosphate Isomerase Terribly Slow rate with Glyceraldehyde…phosphate important in stabilizing binding. Rate Enhancement Due to Proximity (Entropy Reduction) Acid/Base Catalysis Catalytic Mechanisms – acid-base catalysis: give and take protons – covalent catalysis: change reaction paths – metal ion catalysis: use redox cofactors, pKa shifters – electrostatic catalysis: preferential interactions with Transition State. Acid Base Catalysis – Involve Proteins R groups Formation of a Covalent Intermediate Michaelis Menten Curve Michaelis Menten Equation: L. Michaelis and Miss Maud L. Menten. 1913. "Die Kinetik der Invertinwerkung" Biochemische Zeitschrift Vol. 49. vo = Vmax [S] Km + [S] Invertase Reaction: sucrose + H2O glucose + fructose Michaelis Menten Experiment Measure Rate (v) at several concentrations of Substrate (S) Here is one tube with one beginning concentration of S E SP Calculate Δ[S]/min or Δ[P]/min. This enzyme, triosephosphate isomerase is a one substrate, one product enzyme. Michaelis Menten Experiment: Real Data At each [S], the concentration of enzyme is exactly the SAME. Calculate Δ[S]/min for each [S] EOC Problem 6 is about using 340nm light to measure dehydrogenase reactions…the classic lactate dehydrogenase. Do this at more concentrations of S to get a larger data set used for Initial Velocites are the Dashed Line A Michaelis Menten Plot Michaelis Menten Equation is Non-Linear vo = Vmax [S] KM + [S] Straightened Out by reciprocals…Lineweaver Burke Equation: 1/vo = (KM/Vmax)(1/[S]) + 1/Vmax the Equation of a Straight Line y = mx + b Thus, y = 1/vo , x = 1/[S] and m (the slope) = KM/Vmax Lets Plot this Out…next slide Lineweaver-Burke Plot Double Reciprocal Data Points are in this range Origin is Zero There Are Other Equations to Convert the Michaelis Menten Equation to a Straight Line Eadie Hofstie v = -Km(v/S) + Vmax Hanes Wolf: S/v = (1/Vmax)(S) + Km/Vmax all are y = mx + b KM = is an Intrinsic Property of an enzyme What does this mean? Intrinsic vs Extrinsic? Vmax is an Extrinsic Property of Enzymes At a high [S], varying only the enzyme conc : To get an Intrinsic Catalytic Constant from Vmax kcat comes from Vmax and [Enz] Vmax is [molar]/sec [Enz] in molar kcat = Vmax/ [Enz] kcat/Km Calculation of Km and Vmax The enzyme, Practicase Studentose Productate Studentose, mM velocity, μmoles/ml/sec 1 12 2 20 4 29 8 35 12 40 Assay volume = 1 ml/tube What’s in the tube: buffer + enzyme, then add substrate at time Zero. EOC Problems 11(dead easy to do by inspection) and 13 to do by Lineweaver Burke plot Calculation of Km and Vmax Studentose, mM 1/[S] Velocity, 1/v μmoles/ml/sec 1 1 12 0.083 2 0.5 20 0.050 4 0.25 29 0.034 8 0.125 35 0.029 12 0.083 40 0.025 Now Plot this on Lineweaver Burk Plot….remember Zero is near the middle of the graph! Lineweaver Burke Plot of Practicase 1/ 1 Practicase kcat = an Intrinsic Property In the enzyme assay (one ml), each tube had 10 μg of practicase. The molecular weight of practicase is 20,000 D. Thus, each tube had 10 μg / 20,000 μg/μmole = 0.0005 μmole practicase kcat = Vmax/ [Enz] = (50 μmole/sec)/ 0.0005 μmole = 1 x 105 s-1 Thus one enzyme reaction takes 1/ 1x 105 s-1 = 10-5 sec or 10 μ sec. What is Wrong with this L-B graph? What is Wrong with this L-B graph? Things to Know and Do Before Class 1. Principles of catalysis. 2. Enzyme naming concepts. 3. Intrinsic and Extrinsic values of Enzyme kinetics. 4. Be able to do a Michaelis Menten graph. 5. Be able to do a Lineweaver Burke graph. 6. To do EOC problems 1-6, 11, 13.