script - Armed Forces Pest Management Board
advertisement

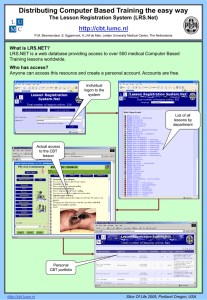
Database Resources of the Defense Pest Management Information Analysis Center Richard G. Robbins, Ph.D. Thank you, LTC Debboun. Hi, everyone. In the brief time we have together this morning, I’d like to introduce you to the databases that are routinely used at DPMIAC to fill literature requests. All are Boolean in nature, and most of you are no doubt already familiar with the first of these, which is our in-house system, the so-called LRS [Slide 1]: http://lrs.afpmb.org/rlgn_app/ar_login/guest/guest Here, in the basic search box, you could enter any search term (authors, keywords, words from the title or text of a paper) and retrieve rafts of hits, but you would almost always be better served by proceeding immediately to our advanced search page [Slide 2]: http://lrs.afpmb.org/BA1G2DVK24Q4KU8A6XFDDQ12/arc/al_03_f0X where, as you can see, you can search by terms in the document title, by author, by terms in the document text, by journal title, and by year range. Let’s work a concrete example. Say I want to search for papers on the ticks of Taiwan. If I specify that the document text must contain all of the words “ticks” and “Taiwan” [Slide 3], I’m going to retrieve an unmanageable number of hits, including many false positives [Slide 4]. However, when we think about it, it’s evident that the focus of our search should be Taiwan, so let’s instead say that “Taiwan” must appear in the document title and “ticks” in the document text [Slide 5]. This search strategy yields a much more manageable list of hits, with virtually no false positives [Slide 6]. A tremendous advantage of the LRS is that all of the documents accessioned here are available as full-text PDFs, which may be viewed on screen, printed, or saved and emailed. Thus, in our last search, if we click on my checklist [Slide 7], we can view it in its entirety and save it wherever we wish [Slide 8]. However, the LRS is not the only database available to DPMIAC customers. Through a contract with the Ovid Corporation, anyone associated with the AFPMB can simultaneously search three powerful biological databases – Agricola, CAB Abstracts, and Medline – and subsequently remove any duplicate hits shared by these databases. Let’s go to the Ovid autologin page [Slide 9]: http://gateway.ovid.com/autologin.cgi Here, by simply typing in our user ID and password (we have only five user ports, so contact me for passwords), we are sent to a page where we are asked to select our databases [Slide 10]. Clicking on Agricola and CAB Abstracts, and then clicking “Continue” [Slide 11], we are sent to our search page, where we can enter the following Boolean search string: (argasid$ or ixodid$ or tick$) and Taiwan [the “$” is a truncation term] which yields a hit list similar to what we saw when working in Advanced mode with the LRS [Slide 12]. We want next to click on “Remove Duplicates” [Slide 13], which prompts a request for deduping preferences [Slide 14]. For the sake of this simple example, we’ll just use the drop-down lists to reverse the positions of the two databases we selected and click on “Continue” [Slide 15]. You see that where hits were shared by our two databases, those in Agricola have been eliminated. We can now view the full text of our hit list, including any available abstracts, and print out, save, or even e-mail this list. Another powerful and popular biological database is Biosis, which is available to DPMIAC customers through a contract with the Dialog Corporation. Let’s go to the Dialog login page [Slide 16]: http://www.dialogclassic.com/ This system differs from Ovid in that our charges are not fixed; rather, using the Dialog databases is like riding in a New York City taxi: you pay for both your hits (your “destination”) and your time online (your time in Midtown traffic). For the sake of the Board’s budget, only DPMIAC personnel run Dialog searches in response to customer requests. Again, let’s try our “ticks of Taiwan” example. First, we type in our user ID and password to get to our search screen [Slide 17], where we type in “b5,” the code for the entire Biosis database, and click on “Submit.” This brings up the boilerplate for Biosis [Slide 18]. We can now enter the following search string: ss(argasid? or ixodid? or tick?) and Taiwan [the “?” is a truncation term] which, as expected, yields a somewhat shorter hit list because we are working with just a single database [Slide 19]. We don’t need to remove duplicates in this case, but we now have many options for viewing the results of our search. One of these involves commands that are summarized by typing in the following: t5/7/all This means that we are asking Dialog to print out the contents of set 5 in option 7 (full bibliographic citation and abstract for each hit), and that we want this process repeated for all the items in our set of search results. Click “Submit” and the entire set appears [Slide 20], to our specifications. We can now view these results, save them, or e-mail them, but whatever we do, it is imperative that we next immediately close out of Dialog because the “taxi meter” is still running. Therefore, on the command line, type “logout” and click “Submit” [Slide 21]. Immediately, the estimated bill for the session is generated and, just to be safe, we can return to the login page and close this browser. Now, you ask, how do I obtain hard copies of papers that I find in my Ovid or Dialog searches but that are not in DPMIAC’s LRS? If the items you seek are biomedical in nature, they can usually be obtained through our DOCLINE contract with the National Library of Medicine. Again, DOCLINE assesses charges on the basis of usage, and the current federal government rate per requested paper is $9. Say, for the sake of argument, that one of my Taiwan tick papers is not in the LRS but, having been found in the Ovid or Dialog databases, we wish to order it. First, we need a PMID (a PubMed identification number), which we can find simply by going to NLM’s PubMed page [Slide 22]: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?tool=LocatorPlusVersionII&holding=nlm lib and typing in suitable search terms, such as “Robbins varanensis.” This yields our PMID: 8691388 [Slide 23]. Now, go to the DOCLINE home page [Slide 24]: http://www.nlm.nih.gov/docline/newdocline.html and select “DOCLINE Login (Registered Users)” [Slide 25]. We enter our user name and password to get to the request page [Slide 26]. Next, click on “Requests” and “Borrow,” which sends us to the Unique Key page [Slide 27], where we type in the PMID (8691388) for the paper we seek. And there it is [Slide 28]. We can now click on “Next,” which will send us to the Routing Instructions page [Slide 29], where I can fill in the data necessary to order a Web PDF of this paper. Web PDFs usually arrive at my desk within 24 hours of my orders; if I order in the morning, I can often expect to receive my PDFs on a same-day basis. For non-biomedical papers, I have stack access at the National Agricultural Library and silk-glove access to the Rare Book Room of the Library of Congress, privileges I’ve enjoyed since 1977. And if even these tools disappoint, well, I defer to my home-town radio station, WQXR: “To be fully informed, read The New York Times.” Thank you, ladies and gentlemen, and sincerest thanks to Mr. David W. Hill for his invaluable assistance in piecing this presentation together.