File - Mr. Holmes Biology
advertisement

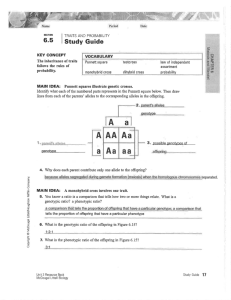
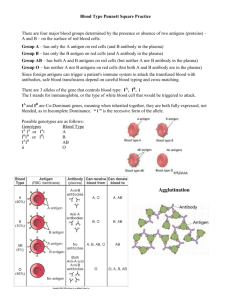
Pea Plant genetics Monohybrid crosses Gregor Mendel discovered genetics through the use of pea plants. One of the more obvious characteristics of pea plants is their flowers. Mendel didn’t know this at first, but purple flowers are the dominant expression(allele) and white flowers are from a recessive allele. P = purple allele p = white allele Parental generation – To create the (P)arental generation, Mendel first spent a great deal of time and effort creating two pure breeding strains of pea plants. One only produced purple flowers and the other only produced white flowers with the respective genotypes of PP and pp. Make a Punnett square for the cross of the parental strains. Also indicate the genotypic and phenotypic ratios. F1 generation – Every offspring produced by crossing Mendel’s parental generation resulted in plants that had purple flowers. White flowers seemed to have disappeared!!! The genotype for all of these plants was Pp. The white flowers were simply masked by the dominant purple allele. When the F1 generation plants are crossed the white flowers come back. Make a Punnett square for the cross of two F1 plants. Include genotypic and phenotypic ratios as well as phenotypic frequencies. F2 generation- The F2 generation had every possible combination of alleles possible for flower color: PP, Pp, and pp. If we were to allow the F2 generation to breed at will with each other, what would be the phenotypic frequencies of their offspring. To get the numbers right, you need to take into account every possible mating situation for each of the F2 genotypes. Some F2 genotypes are more frequent than others. This disparity must be taken into account when calculating overall frequencies. The Punnett square forms on the back of this sheet can help with sorting this out. Dihybrid crosses In his experiments, Mendel discovered a number of traits in pea plants that followed the dominant/recessive pattern that he had found in the flowers. The pods of his pea plants displayed the independent assortment of two of these types of traits: pod shape S-smooth s-constricted and pod color C-yellow c-green. Independent assortment is when one gene does not affect the expression of another gene, even though they may be expressed on the same specific part of the plant/organism(ie: pods). These two traits could be expressed in separate monohybrid Punnett squares of their own, however, since they independently assort, and they both affect the pod, a more comprehensive view of pea plant pods is demonstrated by using a dihybrid Punnett square. Parental generation – In the same vein as the flowers, we would start with two Parent groups of plants that were homozygous dominant for both genes SSCC and another parent group that was homozygous recessive sscc. Make a dihybrid Punnett square for the mating of these two parental strains. Include genotypic and phenotypic ratios. F1 generation – Make a dihybrid square for the crossing of two F1 offspring. Once again, indicate the genotypic ratios, phenotypic ratios, and phenotypic frequencies. F2 generation – There are many different F2 crosses that can be made. Choose two F2 offspring and make a dihybrid square showing the resulting offspring from this mating. Comprehensive Pea Plant genetics As we have learned, the pea plant has many traits that are all expressed at once. For this problem we will be looking at the assortment of these traits into all the frequencies of different offspring that result from a cross between two specific parent individuals. The traits: P-purple flowers /p-white flowers R-round peas /r-wrinkled peas Y-yellow peas /y-green peas S-smooth pod/s-constricted pod C-yellow pod/c-green pod T-tall plant/t-short plant V-vertical flowers/v-lateral flowers PPRrYyssCcTtvv X PprrYySSCcTtVv It is impossible to use a single Punnett square to address this problem . Instead, monohybrid squares or acute observation coupled with the rule of counting will allow you to determine all the possibilities and their frequencies. Show all your work, including monohybrid squares, on a separate sheet of paper. In the end, you do not need to list every possible phenotypic frequency…just the matrix that demonstrates how to calculate each frequency. Parental F1 Genotypic ratio(s) – Phenotypic ratio(s) – F2(x2) F2(x2) Genotypic ratio(s) – Phenotypic ratio(s) – Phenotypic frequencies: F2(x1) F2(x1) Genotypic ratio(s) - Genotypic ratio(s) – Genotypic ratio(s) - Genotypic ratio(s) - Phenotypic ratio(s) – Phenotypic ratio(s) - Phenotypic ratio(s) - Phenotypic frequencies: Phenotypic frequencies: Phenotypic frequencies: Phenotypic ratio(s) Phenotypic frequencies: Combined phenotypic frequency: P F1 Genotypic ratio(s) – Genotypic ratio(s)- Phenotypic ratio(s) – Phenotypic ratio(s)Phenotypic freq. – F2(choice) Comprehensive Workspace Genotypic ratio(s) – Phenotypic ratio(s) – Phenotypic freq. –