Chemical Glycobiology
advertisement

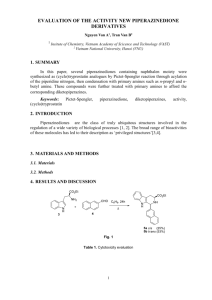
Chemical Glycobiology Carolyn R. Bertozzi1 and Laura L. Kiessling2 Chemical tools have proven indispensable for studies in glycobiology. Synt hetic oligosaccharides and glycoconjugates provide materials for correlating stru cture with function. Synthetic mimics of the complex assemblies found on cell surfaces can modulate cellular interactions and are under development as thera peutic agents. Small molecule inhibitors of carbohydrate biosynthetic and proces sing enzymes can block the assembly of speciÞc oligosaccharide structures. Inh ibitors of carbohydrate recognition and biosynthesis can reveal the biological fu nctions of the carbohydrate epitope and its cognate receptors. Carbohydrate bio synthetic pathways are often amenable to interception with synthetic unnatural s ubstrates. Such metabolic interference can block the expression of oligosacchari des or alter thestructures of the sugars presented on cells. Collectively, these c hemical approaches are contributing great insight into the myriad biological fun ctions of oligosaccharides. Oligosaccharides and glycoconjugates (glycoproteins and glycolipids) have i ntrigued biologists for decades as mediators of complex cellular events. With r espet to structural diversity, they have the capacity to far exceed proteins and nucleic acids. This structural variance allows them to encode information for s pecific molecular recognition and to serve as determinants of protein folding, st ability, and pharmacokinetics. Given that glycosylation is one of the mostubiqui tious forms of posttranslational modification, the unexpectedly small number of genes identified in the initial analyses of the human genome sequence provide s even more impetus for understanding the biological roles of oligosaccharides. Oligosaccharide functions are now being elucidated in molecular detail, but advances in glycobiology have been slow to arrive compared with the pace of revelations in protein or nucleic acid biochemistry. The same structural diversi ty that has captivated biologists has also frustrated efforts to define oligosaccharide expression patterns on proteins and cells and to correlate struct ure with function. Some technical challenges are analytical in nature; determina tion of the oligosaccharide sequence on a specific glycoconjugate is still far fr om routine. Others originate from glycoconjugate biosynthesis, which is neither template-driven nor under direct transcriptional control. Oligosaccharides are as sembled in step-wise fashion primarily in the endoplasmic reticulum and Golgi apparatus (Fig. 1), a process that affords significant product microheterogeneit y(1). As a result, it is difficult to obtain homogeneous homogeneous and chem ically defined glycoconjugates from biological sources. Without such materials n hand, biological functions are difficult to unravel. Genetic approaches have contributed considerably to our appreciation of oli gosaccharide function. The availability of entire genome sequences has revealed the multiplicity of enzymes that contribute to glycoconjugate assembly. Their deletion in model organisms has provided substantial insight. For example, mic e deficient in a mannosidase II expressed an altered portfolio of N-linked glyc ans on their cell surface glycoproteins (2). The mice were prone to a systemic autoimmune response, suggesting that abnormalities in N-glycosylation in hum ans may be a factor in the pathogenesis of autoimmunity. Still, cell surface pr esentation of simple as well as complex glycans requires many genes to be ex pressed in concert, which complicates the analysis of single gene “knockouts” or “knockins.” As outlined in this review, chemical approaches are powerful allies to gen etics and biochemistry in the study of glycobiology. Chemical tools have been used to probe glycosylation at many levels. For example, cell surface carbohyd rate-receptor binding events (Fig. 1) can be interrogated with synthetic oligosac charides, glycoconjugates, and their analogs. The biosynthesis of oligosaccharide structures can be disrupted or modulated by synthetic enzyme inhibitors. Unnat ural sugars that substitute for native monosaccharides or their downstream meta bolic intermediates Fig. 1. Glycoconjugate biosynthesis and cell surface recognition. Exogenously s uppliedmonosaccharides are taken up by cells and converted to monosaccharide Òbuilding blocksÓ (typically nucleoside sugars) inside the cell. Several steps of metabolic transformation might take place en route from an exogenous sugar to a building block. The building blocks are imported into the secretory comp artments where they are assembled by glycosyltransferases into oligosaccharides bound to a protein (or lipid) scaffold. In the case of N-linked glycoproteins, a core oligosaccharide is assembled in the cytosol, then transported into the ER where it is processed by glycosidases, and then further elaborated by glycosyl transferases. Once expressed in fully mature form on the cell surface, the glyc oconjugates can serve as ligands for receptors on other cells or pathogens. Che mical tools can be used to inhibit or control any stage of this process. (Fig. 1) can intercept biosynthetic pathways, leading to changes in cell surface glycosylation. These chemical strategies allow one to perturb glycosylation and oligosaccharide-receptor interactions in a cellular context. Furthermore, chemically synthesized molecules that disrupt pathological carbohydrate-dependent processes are emerging as important therapeutic agents. Synthesis of Oligosaccharides and Glycoproteins Access to structurally defined oligosaccharides and glycoconjugates is a prerequisite for unraveling their function. Chemical routes to the production of oligosaccharides are, therefore, essential. Advances on this front are providing materials for the assessment of glycan function, the establishment of the structural features important for function, the elucidation of biosynthetic pathways, the creation of carbohydrate-based vaccines, the production of non-natural glycosylated antibiotics, and the generation of inhibitors of glycoconjugate function. Two general strategies are used for in vitro oligosaccharide production: enzymatic(including chemoenzymatic) synthesis and chemical synthesis. In enzymatic and chemoenzymatic routes, saccharide intermediates are elaborated with enzymes, typically glycosyltransferases or glycosidases, to generate oligosaccharides (Fig. 2A) (3–6). Chemoenzymatic synthesis is distinguished from enzymatic synthesis by its reliance on both chemical synthetic and enzymatic transformations. In chemical synthesis, the appropri appropriate building blocks are produced and assembled into oligosaccharides (Fig. 2B). In both approaches, the focus is on forming the critical connection that links saccharide building blocks: the glycosidic bond. Chemical synthesis and enzyme-based routes are complementary. Enzymes can be used to effect glycosylation with absolute regio- and stereo-control. If the necessary enzyme is available, the desired bond can be formed (Fig. 2A), often with high efficiency. In comparison, chemical synthesis offers exceptional flexibility. Natural and non-natural saccharide building blocks can be assembled with natural or non-natural linkages. Although some enzymes will act on alternative substrates, chemical synthesis provides the means to generate any oligosaccharide, oligosaccharide analog, or glycoconjugate. The chemical synthesis of oligosaccharides is formidable. It requires stereochemical and regiochemical control in glycosidic linkage formation. The first viable method for controlled glycosidic bond formation, the Koenigs-Knorr glycosylation, was reported in 1901 (7). Although the search for alternative glycosylation reactions is ongoing (8), chemists have made remarkable strides in carbohydrate synthesis. The problem of obtaining the proper regiochemistry of glycosylation has been largely solved by the development of orthogonal hydroxyl group protection strategies. Thus, groups can be installed to block reaction at one site and later removed to unmask specific hydroxyl groups for glycosylation. To devise safer and less toxic procedures, reactions that require heavy Fig. 2. Generic examples of glycosylation reactions. (A) Enzyme-catalyzed glycosylation. Glycosyltransferases catalyze the formation of glycosyl bonds from sugar nucleotide donors. The speciÞc regioisomer and stereoisomer produced depends on the enzyme employed. (B) Chemical glycosylation. Most such glycosylations involve the reaction of a glycosyl donor, equipped with a leaving group (X) at the anomeric position, with a glycosyl acceptor. To produce the desired regioisomer, protecting groups (P9,P) are used to block other reactive sites. The desired stereoisomer can be generated by altering the nature of X or the protecting group. metal activators are being replaced with milder, more environmentally sound methods (9, 10). Our understanding of how to obtain the desired configuration of a glycosidic bond is also more sophisticated. Stereochemical control can be achieved by employing stereospecific activation methods, using protecting groups that direct the orientation of the glycosidic bond through intermolecular (neighboring group participation) or intramolecular (tethered aglycone delivery) participation, altering the steric environment around the anomeric position to bias the desired outcome, or exploiting the intrinsic stereoelectronic preferences for reaction at the anomeric position. Two major advances are being applied to streamline the chemical synthesi s of oligosaccharides: one-pot reactions (11, 12) and polymer-supported synthesi s (13, 14). In one-pot processes, glycosylation reactions occur sequentially in a single reaction vessel; the most reactive glycosyl donor is triggered first and t he least is coerced to engage in the final reaction. A key concept underlying p rogress on this front is that there are “armed” (reactive) and “disarmed” (less r eactive) glycosyl donors (10, 15). A number of research groups have exploitedt his knowledge to efficiently assemble oligosaccharides using one-pot reactions (12, 16–20). Solidphase synthesis of oligosaccharides similarly offers powerful advantages for oligosaccharide synthesis in comparison to conventional methods because it circumvents multiple purification steps needed in traditional solution syntheses. Pioneering studies in solid-phaseoligosaccharide synthesis were initiat ed in the early 1970s (21) after Merrifield’s successful demonstration of solid-p hase peptide synthesis (22). Unfortunately, the harsh conditions needed for som e of the early glycosylation reactions and the difficulty of monitoring the reacti on on the solid support hindered progress for 20 years. Newly developed glyco sylation methods and advances in solid-phase organic chemistry rejuvenated inte rest in solid-phase oligosaccharide synthesis in the 1990s (13, 14). The product ivity from these renewed efforts is evident from the diversity of and complexit y of oligosaccharides that have been synthesized, including sequences containin g up to 12 residues (23). Progress on this front also has facilitated the generat ion of oligosaccharide libraries containing up to 1300 compounds (23–25). Thu s, complex oligosaccharides and oligosaccharide libraries are becoming accessibl e to a large community of scientists. Most oligosaccharides are linked covalently to proteins. As oligosaccharides become more accessible, the next challenge is to integrate them into glycoproteins. Several laboratories have used protected glycosylated amino acids as building blocks for automated solid-phase peptide synthesis (SPPS) to gen- erate glycosylated peptide fragments reminiscent of natural glycoproteins (26). Modern FMOC-based peptide synthesis methods are sufficiently mild that the oligosaccharide remains intact throughout the synthesis. As an illustration, Danishefsky and co-workers chemically synthesized glycopeptide 2, derived from the mucin-like leukocyte antigen leukosialin (CD43), using trisaccharide–amino acid 1 as a building block (Fig. 3A) (27). The oligosaccharide structures and their arrangement on the polypeptide backbone are characteristic of tumor-associated glycoproteins and, hence, such synthetic assemblies may serve as tumor vaccine components. The extension of these methods to fulllength glycoproteins has proved more troublesome, largely due to limitations inherent to linear, step-wise SPPS. Peptides larger than 50 to 60 residues are difficult to obtain using conventional methods due to poor yields and accumulating byproducts. Much larger polypeptides can be produced using recombinant DNA technology; thus, several groups have exploited recombinant proteins as starting materials for the synthesis (i.e., “semi-synthesis”) of homogeneous glycoproteins. Whereas the glycosidic linkage is too difficult a bond to be made between an oligosaccharide and a large protein, analogs of glycoproteins bearing unnatural linkages are readily prepared (28). These “neoglycoproteins” lend themselves to a facile convergent assembly from proteins and synthetic oligosaccharides oligosaccharides. But, their non-native linkages may affect their overall structures and perturb their biological activities. The total chemical synthesis of native glycoproteins has recently been facilitated by breakthroughs in protein chemistry. In particular, the “native chemical ligation” method (29, 30) has enabled the convergent condensation of unprotected glycopeptide fragments, each generated by automated SPPS, to form full-length, fully functional glycoproteins (31) (Fig. 3B). The related “expressed protein ligation” (32) permits the chemical union of recombinant protein fragments with synthetic glycopeptides, further relieving the burden of large protein synthesis. O-linked (33) and N-linked (34) glycoproteins bearing homogeneous and chemically defined glycans have been prepared in this fashion, suggesting that any glycoprotein may now be obtained in the quantities required for structure determination and functional analysis. These synthetic advances are very important with respect to development of glycoprotein therapeutics. Glycosylated biotechnology products such as monoclonal antibodies, erythropoietin, and tissue plasminogen activator may benefit from methods for their semi-synthesis. What future breakthroughs can we expect in the synthesis of oligosaccharides and glycoconjugates? Success in the automated solid- phase synthesis of complex oligosaccharides appears imminent (35) and efforts to automate one-pot assembly methods are advancing These developments offer nonspecialists access to oligosaccharides that can be used to address problems in biology. Although a standard method for the synthesis of oligosaccharides has yet to emerge, the diversity of glycosidic linkages may demand multiple synthetic approaches. Applications of these methods will afford diverse combinatorial libraries of oligosaccharides and more complex glycoconjugates. This increased repertoire of compounds will facilitate the identification of specifi oligosaccharide and glycoprotein ligands for proteins and provide new leads for inhibitor design. Inhibitors of Glycan Biosynthesis and Processing The discovery of diverse biological roles for oligosaccharides and glycoconjugates is fueling interest in the development of chemical tools that block their formation and/or function. Two general types of inhibitors are being sought: those that block glycoconjugate biosynthesis and those that interfere with glycoconjugate recognition. Effective inhibitors of various biosynthetic steps in glycoconjugate assembly have the potential to transform our understanding of carbohydrate function. By blocking the production of specific glycoconjugates, their biological roles can be ascertained (36). Similarly, antagonists that prevent glycoconjugate recognition can illuminate the function of the natural interactions (37). Progress on all of these fronts is accelerating. Fig. 3. Chemical synthesis of glycopeptides and glycoproteins. (A) Glycopeptide synthesis using glycosylated amino acid building blocks. Synthesis of a mucin-like fragment of leukosialic (CD43) (2) by incorporation of glycosylated FMOC amino acids (1) into solid-phase peptide synthesis (SPPS). (B) Glycoprotein synthesis by convergent coupling of glycopeptide fragments. Assembly from synthetic glycopeptide fragments using the technique of native chemical ligation. One fragment is functionalized as a COOH-terminal thioester (athioester), and the other bears an NH2-terminal cysteine residue. A transthioesteriÞcation reaction between the two components produces an intermediate thioester that rearranges to the peptide bond shown in the product. Efforts to generate antagonists of the biosynthetic and processing enzymes have been successful (38–40). To produce oligosaccharides, eukaryotic organisms require enzymes for both synthesis and remodeling. The former is mediated by glycosyltransferases, catalysts that mediate the formation of glycosidic bonds, and the latter by glycosidases, enzymes that hydrolyze glycosidic bonds. The glycosidases have proven to be particularly vulnerable targets for inhibition. Natural product inhibitors have been identified, and numerous antagonists inspired by these have been synthesized (41, 42). In a notable example of computer-aided design, potent transition state inhibitors of influenza virus N-acetylneuraminidase were devised (40, 43). These agents, which interfere with viral infectivity, are currently on the market. Several inhibitors of another major class of carbohydrate-modifying enzymes, the glycosyltransferases, have been reported. Compound 3 (Fig. 4), for example, is a potent inhibitor of an a-2,6-sialyltransferase that uses cytidine monophosphate (CMP)–sialic acid as a glycosyl donor substrate (44). Although active against the enzyme, polar and charged compounds of this type are unlikely to be effective in cells or organisms due to their membrane impermeability. Thus, strategies for the design and discovery of glycosyltransferase inhibitors that can access their targets inside cells are needed. The oligosaccharyl transferase inhibitor 4 (Fig. 4) is capable of transport across the endoplasmic reticulum membrane where it can act on its target and abrogate N-linked glycosylation (45), and other glyc osyltransferase inhibitors demonstrate efficacy in whole cells (38). Library scree ning approaches that are now routine in pharmaceutical industry might be appli ed to the glycosyltransferases (46). The availability of combinatorial methods f or synthesizing libraries of drug-like (and, hence, bioavailable) compounds has revolutionized the search for pharmacological tools in academic laboratories. In deed, library screens have produced cell-permeable, small molecule inhibitors of a class of carbohydrate modifying enzymes called sulfotransferases (5, Fig. 4), and these may be effective tools for delineating biological function (47). The prospects for advances in glycosyltransferase inhibitor identification are, therefor e,excellent. In addition, the rapid increase in structural information available for this class of proteins willaid in inhibitor design and discovery (48, 49). Inhibitors of Glycan Recognition The generation of compounds that block glycan recognition remains a major challenge. Many oligosaccharide binding sites are relatively shallow and solvent exposed, and binding interactions can take place over large surface areas (50). Those confronted with the problem of inhibiting protein–saccharide complexation encounter many of the same challenges as those seeking to block protein–protein interactions. Moreover, in solution many oligosaccharides bind their protein targets with relatively low affinities (e.g., with adissociation constant Kd ' 1023 to 1024 M);thus, initial lead compounds tend to require much optimization. Lastly, structural data is often lacking, so information on the important binding contacts can be obtained only through the synthesis and evaluation of analogs. Given these barriers, the progress that has been made in identifying inhibitors of one family of carbohydrate-binding proteins, the selectins, is impressive. The selectins are a family of three carbohydrate-binding proteins that have been the object of numerous investigations because of their role in leukocyte recruitment to sites of inflammation (51). The discovery of the selectins led to the initial identification of a ligand, the tetrasaccharide sialyl Lewis x (sLex). The findings raised questions illustrative of those generally relevant for protein – carbohydrate interacinteractions: What functional groups are needed for recog nition by the selectins? Can thesLex tetrasaccharide be modified such that it bi nds specific selectin family members? Can compounds that bind more tightly b e discovered? Access to sLex and a wide array of analogs, conjugates, and mi mics has been instrumental in addressing these questions. Data from many synthetic sLex derivatives have revealed the critical contacts for selectin binding, information that has guided the generation of more potent inhibitors. The functional groups important for binding to each of the selectin family members (E-, Pand L-selectin) have been identified. For instance, the hypothesis that sulfation of sLex would increase its affinity for L-selectin (52) was confirmed with derivatives produced by chemical synthesis (53–55). Similarly, a chemoenzymatic route was used to generatesLex-substituted glycopeptides to elucidate the critical binding epitope of PSGL-1 (56), the physiological ligand for P-selectin. With knowledge of the sLex functional groups important for complexation, mimics of the tetrasaccharide have been synthesized that are potent selectin antagonists (57). For example, tetrasaccharide mimic 6 (Fig. 5) is .50-fold more active at blocking E-selectin than sLex (58). In a P-selectin inhibition assay, macrocyclicsLex analog 7 exhibits a dramatic enhancement (about 103) relative to sLex (59). These data underscore the tremendous progress made in generating efficacious glycomimetics. Synthesis and Biological Activity of Glycoassemblies The function of many carbohydrates is contingent on their multidentate presentation. The binding of proteins to monovalent carbohydrate determinants is often weak, yet the strength and specificity required for recognition in physiological settings is high. The simultaneous formation of multiple protein–carbohydrate interactions is one binding mode that can be exploited to achieve the necessary avidity. In physiological settings, saccharide epitopes and their protein receptors are arranged such that multiple binding events can occur simultaneously. Naturally occurring carbohydrate displays are widespread: examples include highly glycosylated proteins (e.g., mucins), the carbohydrate surfaces of bacteria and other pathogens, and the outer membranes of mammalian cells. Carbohydrate-binding proteins also tend to be oligomeric or present in multiple copies at the surface of a cell. The interaction of multivalent presentations can resultin the formation of numerous simultaneous complexation events that proceed to afford a high observed affinity and a high functional affinity (60). What are the physiological advantages conferred by multivalent binding? Synthetic arrays have provided key answers to this question. First, these interactions have been shown to be highly specific and versatile (61–63). For example, binding can be modulated by changing the individual saccharide residues or by altering their spacing. Second, the kinetics exhibited by such binding events are likely critical for biological systems. For example, relative to monovalent binding, multivalent interactions exhibit greater reversibility in the presence of competing ligands (64). Thus, low affinity, multivalent interactions are less likely to entrap cells in unproductive binding events. In addition, binding events mediated by multiple weak interactions are expected to be more resistant to shear stress, such as that encountered when cells interact in the bloodstream (65). To understand the roles of multivalency in carbohydrate recognition, platforms that display multiple copies of recognition elements have been generated (66–68). A number of diverse scaffolds have been used for multivalent presentation; these include low–molecular weight displays (e.g., dimers and trimers), dendrimers, polymers, and liposomes. The structure of the display determines what features of selected of naturally occurring multivalent ligands it mimics. For example, a low–molecular weight ligand or dendrimer can resemble a branched oligosaccharide chain, such as those displayed by glycoproteins. Alternatively, larger displays such as polymers or liposomes can more effectively mimic a glycoprotein or a glycolipid array. There are several distinct mechanisms that contribute to the high activities often observed for multivalent ligands. An understanding of these is critical for optimizing ligand performance and for understanding how natural systems function. Relevant mechanisms include the chelate effect, occupation of adjacent subsites, and ligand-induced protein clustering. When the chelate effect operates (Fig. 6A), multiple interactions occur after formation of the first contact; these are facilitated because of the high effective concentration of the binding groups (69). This mode of binding can give rise to large enhancements in activity if the orientation and display of binding groups are favorably disposed. Because multipoint binding typically involves ligand organization, it exacts a conformational entropy penalty that may offset the advantages of chelation. Alternatively, multivalent displays may be effective ligands because they occupy subsites as well as the primary binding site on the target protein. Many lectins possess secondary sites adjacent the key binding cleft (Fig. 6B) (50). Lastly, the activities of many small and large multivalent carbohydrate derivatives are due to their abilities to cluster theirreceptors (Fig. 6C) (70, 71). Interestingly, ligands with this property may serve not only as antagonists but also as agonists. Because cell surface receptor clustering can facilitate signaling transduction, carbohydrate displays that cluster receptors or lectins that cluster glycoproteins can elicit cellular responses. Thus, multivalent ligands can exhibit a wide range of different activities, depending on their binding modes. Two recent studies dramatically illustrate the power of multivalent presentation in inhibitor design and the interplay of different binding mechanisms. Highly potent pentavalent inhibitors of bacterial toxin binding to host cells were synthesized. The toxins targeted, heat-labile enterotoxin (LT-1) and shiga-like toxin I (SLT-I), are members of the class of AB5 toxins. The AB5 class, which can be subdivided into the cholera, pertussis, and shiga toxin families, possess a pentagonal arrangement of five B subunits and a single A subunit (72). These toxins, which are responsible for millions of human fatalities each year (73), invade cells by multivalent binding of the B subunit to the carbohydrate residues of gangliosides. Thus, a logical strategy is to generate multivalent ligands that can occupy all five binding sites simultaneously. Fan et al. applied this strategy to produce candidate inhibitors of LT-1, which were generated by appending galactose residues to a pentacyclen core (Fig. 7) (74). A potent antagonist of LT-1 was found, a compound 105 times more active than the corresponding monovalent galactose derivative. This excellent potency appears to arise from its ability to interact simultaneously with the five toxin B subunits. Kitov et al. pursued an alternative strategy for inhibiting SLT-I (75). Structural studies of SLT-I complexed with the glycolipid Gb3 revealed that each B subunit possessed subsites adjacent to the primary carbohydrate-binding site. They synthesized dimers of the appropriate trisaccharide binding element that could, in principle, occupy both the primary and secondary site. Although their divalent ligands exhibited only modest increases in potency (40-fold), attachment of these units to a pentavalent scaffold afforded one of the most potent (107times more active than the trisaccharide alone) multivalent ligands (8, Fig. 7) yet described. Structural analysis of compound 8 bound to SLT-1 did not reveal the 1:1 complex envisioned but rather a complex containing two equivalents of the pentameric protein. This ligand appears to act both through dimerizing the target receptor as well as through the chelate effect. The excellent potencies of the toxin inhibitors highlight the advantages of multivalent presentation, and the wide range of unique binding modes multivalent ligands can employ. Multivalent presentation is also beneficial for eliciting rather than inhibiting biological responses. This role has long been appreciated in the context of vaccine development. As early as 1929 (76), it was recognized that displays of oligosaccharides conjugated to proteins elicit the production of specific antibodies to carbohydrate. In 1975, Lemieux and co-workers showed that defined synthetic glycoconjugates could be synthesized and used to elicit an immune response (77). These investigations provided the foundation Fig. 5 (left). Examples of selectin antagonists (6 and 7). Both were designed to mimic the tetrasaccharide sLex. Fig. 6 (right). Binding modes engaged in by multivalent ligands. (A) Multivalent ligands can bind oligomeric receptors by occupying multiple binding sites (chelate effect), (B) occupying primary and secondary binding sites on a receptor, and (C) causing receptor clustering in the membrane. Fig. 7 (left). Multivalent ligands can act as antagonists and agonists of biological processes. Compound 8 is a potent antagonist of shiga-like toxin I. Multivalent ligand 9 elicits the downregulation of L-selectin, thereby inhibiting L-selectin function. Fig. 8 (right). Modulating cell surface glycosylation by metabolic interference. (A) Unnatural substrates fed to cells can divert oligosaccharide biosynthesis away from endogenous scaffolds, reducing the expression of speciÞc carbohydrate structures. (B) Unnatural substrates can be used in biosynthetic pathways and incorporated into cell surface glycoconjugates. (C) If the unnatural substrates possess unique functional groups, their metabolic products on the cell surface can be chemically elaborated with exogenous reagents(D). for current approaches to generating synthetic carbohydrate-based vaccines. One such effort is directed at exploring the features required to generate the first vaccine against Shigella dysenteriae (78). Synthetic anti-cancer vaccines are also being pursued. Danishefsky and co-workers have used state-of-the-art synthetic methods to generate complex glycoconjugates (79) with promising anti-cancer activities. To optimize the potency of carbohydrate- based vaccines, more data addressing how the structure of the conjugate (epitopespacing, valency, backbone composition) affects its immunogenicity is needed. Multivalent ligands can also act as effectors of one response and inhibitors of another. In one study, a multivalent saccharide derivative was designed to effect aprocess that disables the receptor, L-selectin (80, 81). L-Selectin is a transmembrane protein found on neutrophils and lymphocytes that facilitates the transient attachment and rolling of leukocytes through its interaction with highly O-glycosylated proproteins on the endothelium (82). A soluble form of L-selectin can be released into circulation by a membrane-associated protease (83). Given the role of cell surface L-selectin, ligands that induce its downregulation could serve as anti-inflammatory agents. To test this hypothesis, human neutrophils were treated with monovalent and multivalent oligosaccharides that mimic features of physiologic L-selectin ligands. The levels of L-selectin after addition of monovalent oligosaccharides were unchanged, but upon exposure to multivalent display 9 (Fig. 7) L-selectin was lost from the surface (80, 81). Multivalent ligand 9 also inhibits L-selectin-mediated cell rolling, suggesting that its ability to downregulate L-selectin makes it a highly effective inhibitor (84). These results suggest a new approach to manipulating the cell surface interactions. The design of multivalent ligands that selectively disarm receptors by activating endogenous processes is an uncharted territory with high potential. Modulating Cell Surface Glycosylation by Metabolic Interference The ability to alter glycoconjugate structures expressed on cell surfaces is important for understanding their biological functions. An alternative to enzyme inhibitors is the use of unnatural metabolic substrates that can intercept carbohydrate biosynthetic pathways (85). Metabolic interference can produce several outcomes on the cell surface (Fig. 8). An unnatural substrate might divert oligosaccharide elaboration away from endogenous scaffolds destined for the cell surface (Fig. 8A). The result is a reduction in the amount of mature structures expressed by the cell. Alternatively, unnatural substrates might be designed to engage a biosynthetic pathway, resulting in their incorporation into cell surface glycoconjugates (Fig. 8B). The result is the presentation of an unnatural epitope that might display different receptor binding properties than its native counterpart. Subtle modifications to the fine structure of a monosaccharide can be engendered in this fashion. Incorporation of unnatural sugars with reactive functional groups into cell surface glycoconjugates (Fig. 8C) provides a scenario in which the glycan structure can be further altered by chemical reactions at the cell surface (Fig. 8D). An example of cell surface glycan suppression using metabolic decoys is provided by the “oligosaccharide primers” described by Esko and co-workers. When fed to cells, hydrophobic disaccharides intercepted the biosynthesis of sLex, and the cells displayed a concomitant reduction in cell surface E-selectin binding activity (86). The roles of sLex in inflammation and tumor metastasis may be addressed using this reversible process. Likewise, hydrophobic glycosides of xylose can distract the glycosaminoglycan biosynthetic machinery, thereby temporarily reducing the density of these chains on cell surfaces (87). The pathway for sialic acid biosynthesis is highly amenable to biosynthetic modulation using unnatural metabolic precursors. Derived from metabolism of N-acetylmannosamine (ManNAc) (10, Fig. 9), sialic acids are known to participate in myriad cell surface recognition events, including selectinmediated leukocyte adhesion and influenza virus binding. Synthetic analogs of ManNAc bearing unnatural N-acyl groups are substratesfor the metabolic pathway, and when fed to cells they produce unnatural sialic acids on cell surface glycoconjugates (Fig. 9) (88). Cellular expression of unnatural sialic acids can either inhibit or enhance viral infection, depending on the physical interaction of the unnatural moiety with the viral receptor (89, 90). Unnatural sialosides can also disrupt contact inhibition of cell growth (91), and block the binding of myelin-associated glycoprotein to neurons (92). The effects witnessed implicate sialic acid residues as key determinants of the respective processes. If the unnatural N-acyl substituent comprises a unique chemical group such as a ketone (93) or azide (94), chemoselective reactions can be performed on the cells that further augment their cell surface composition. This technique has been used to construct nonnatural glycans on cell surfaces for lectin binding studies (95) and to target magnetic resonance imaging contrast reagents to cells overexpressing sialic acid residues, a hallmark of many tumor types (96). Unnatural sialic acids bearing ketone groups have also been used to facilitate viral-mediated gene delivery (97). Other sugars such as N-acetylgalactosamine (GalNAc) can also be replaced by unnatural variants using metabolic processes (98). If the approach is generalizable across carbohydrate biosynthetic pathways, multiple modifications might be made to cell surface glycans for structure/function studies in a cellular context. In summary, advances in oligosaccharide and glycoprotein synthesis have provided the critical material for biological investigations. Genome sequencing efforts have unveiled the various enzymes that participate in glycoconjugate biosynthesis and processing, and a detailed understanding of these pathways will allow the judicious choice of targets for inhibitor design and metabolic interference. The convergence of chemical tools with frontier genetic and biochemical technologies has created an exciting platform from which to tackle problems in glycobiology References and Notes 1. N. Jenkins, R. B. Parekh, D. C. James, Nature Biotechnol.14, 975 (1996). 2. D. Chui et al., Proc. Natl. Acad. Sci. U.S.A. 98, 1142(2001). 3. S. L. Flitsch, Curr. Opin. Chem. Biol. 4, 619 (2000). 4. M. M. Palcic, Curr. Opin. Biotechnol. 10, 616 (1999) 5. S. David, C. Auge, C. Gautheron, Adv. Carbohydr.Chem. Biochem. 49, 175 (1991). 6. E. J. Toone, E. S. Simon, M. D. Bednarski, G. M.Whitesides, Tetrahedron 45, 5365 (1989). 7. W. Koenigs, E. Knorr, Berichte 34, 957 (1901). 8. S. H. Khan, R. A. OÕNeill, Eds., Modern Methods inCarbohydrate Synthesis; Frontiers in Natural ProductResearch (Harwood Academic, Australia, 1996), vol.1; Preparative Carbohydrate Chemistry (Dekker, NewYork, 1997). 9. B. G. Davis, J. Chem. Soc. Perkin Trans. I, 2137 (2000). 10. H. Paulsen, Angew. Chem. Int. Ed. 21, 155 (1982). 11. K. M. Koeller, C. H. Wong, Chem. Rev. 100, 4465(2000). 12. N. L. Douglas, S. V. Ley, U. Lu¬cking, S. L. Warriner,J. Chem. Soc., Perkin Trans. I, 51 (1998). 13. P. H. Seeberger, W.-C. Haase, Chem. Rev. 100, 4549(2000). 14. D. Kahne, Curr. Opin. Chem. Biol. 1, 130 (1997). 15. B. Fraser-Reid, D. R. Mootoo, P. Konradsson, U. Udodong,J. Am. Chem. Soc. 110, 5583 (1988). 16. Z. Zhang et al., J. Am. Chem. Soc. 121, 734 (1999). 17. J. Gildersleeve, A. Smith, K. Sakurai, S. Raghavan, D.Kahne, J. Am. Chem. Soc. 121, 6176 (1999). 18. R. Geurtsen, D. S. Holmes, G. J. Boons, J. Org. Chem.62, 8145 (1997). 19. H. Yamada, T. Harada, T. Takahashi, J. Am. Chem.Soc. 116, 7919 (1994). 20. S. Raghavan, D. Kahne, J. Am. Chem. Soc. 115, 1580(1993). 21. J. M. Fre«chet, in Polymer-Supported Reactions in OrganicSynthesis P. Hodge, D. C. Sherrington, Eds.(Wiley, Chichester, UK, 1980), pp. 407Ð434. 22. R. B. MerriÞeld, J. Am. Chem. Soc. 85, 2149 (1963). 23. K. C. Nicolaou, N. Winssinger, J. Pastor, F. DeRoose,J. Am. Chem. Soc. 119, 449 (1997). 24. R. Liang et al., Science 274, 1520 (1996). 25. M. J. SoÞa et al., J. Med. Chem. 42, 3193 (1999). 26. H. Herzner, T, Reipen, M. Schultz, H. Kunz, Chem. Rev.100, 4495 (2000). 27. D. Sames, X.-T. Chen, S. J. Danishefsky, Nature 389,587 (1997). 28. L. Marcaurelle, C. R. Bertozzi, Chem. Eur. J. 5, 1384(1999). 29. P. E. Dawson, T. W. Muir, I. Clark-Lewis, S. B. H. Kent,Science 266, 776 (1994). 30. G. J. Cotton,T. W. Muir, Chem. Biol. 6, R247 (1999). 31. Y. Shin et al., J. Am. Chem. Soc. 121, 11684 (1999). 32. T. W. Muir, D. Sondhi, P. A. Cole, Proc. Natl. Acad. Sci.U.S.A. 95, 6705 (1998). 33. D. Macmillan, C. R. Bertozzi, Tetrahedron 56, 9515(2000). 34. T. J. Tolbert, C.-H. Wong, J. Am. Chem. Soc. 122,5421 (2000). 35. O. J. Plante, E. R. Palmacci, P. H. Seeberger, Science 291,1523 (2001). 36. A. D. Elbein, Annu. Rev. Biochem. 56, 497 (1987). 37. P. Sears, C. H. Wong, Angew. Chem. Int. Ed. 38, 2300(1999). 38. F. M. Platt et al., Science 276, 428 (1997). 39. Y. J. Kim, M. Ichikawa, Y. Ichikawa, J. Am. Chem. Soc.121, 5829 (1999). 40. M. von Itzstein et al., Nature 363, 418 (1993). 41. A. Berecibar, C. Grandjean, A. Siriwardena, Chem. Rev.99, 779 (1999). 42. B. Ganem, Acc. Chem. Res. 29, 340 (1996). 43. C. U. Kim et al., J. Am. Chem. Soc. 119, 681 (1997). 44. B. Mu¬ller, C. Schaub, R. R. Schmidt, Angew. Chem. Int.Ed. 37, 2893 (1998). 45. P. D. Eason, B. Imperiali, Biochemistry 38, 5430 (1999). 46. G. Jung, Ed., Combinatorial Chemistry: Synthesis,Analysis, Screening (Wiley-VCH, Weinheim, Germany,1999). 47. J. I. Armstrong et al., Angew. Chem. Int. Ed., 39, 1303(2000). 48. U. M. Unligil, J. M. Rini, Curr. Opin. Struct. Biol. 10,510 (2000). 49. C. Breton, A. Imberty, Curr. Opin. Struct. Biol. 9, 563(1999). 50. W. I. Weis, K. Drickamer, Annu. Rev. Biochem. 65, 441(1996). 51. L. A. Lasky, Annu. Rev. Biochem. 64, 113 (1995). 52. S. Hemmerich, H. Lefßer, S. D. Rosen, J. Biol. Chem.270, 12035 (1995). 53. C. Galustrian et al., Biochem. Biophys. Res. Commun.240, 748 (1997). 54. C. R. Bertozzi, S. Fukuda, S. D. Rosen, Biochemistry 34,14271 (1995). 55. W. J. Sanders, T. R. Katsumoto, C. R. Bertozzi, S. D.Rosen, L. L. Kiessling, Biochemistry 35, 14862 (1996). 56. A. Leppanen, S. P. White, J. Helin, R. P. McEver, R. D.Cummings, J. Biol. Chem. 275, 39569 (2000). 57. E. E. Simanek, G. J. McGarvey, J. A. Jablonowski, C.-H.Wong, Chem. Rev. 98, 833 (1998). 58. R. Banteli et al., Helv. Chim. Acta 83, 2893 (2000). 59. C.-Y. Tsai, X. Huang, C.-H. Wong, Tetrahedron Lett.41, 9499 (2000). 60. M. Mammen, S.-K. Choi, G. M. Whitesides, Angew.Chem. Int. Ed. 37, 2754 (1998). 61. K. H. Mortell, R. V. Weatherman, L. L. Kiessling, J. Am.Chem. Soc. 118, 2297 (1996). 62. R. Liang et al., Proc. Natl. Acad. Sci. U.S.A. 94, 10554(1997). 63. R. V. Weatherman, K. H. Mortell, M. Chervenak, L. L.Kiessling, E. J. Toone, Biochemistry 35, 3619 (1996). 64. J. H. Rao, J. Lahiri, L. Isaacs, R. M. Weis, G. M. Whitesides,Science 280, 708 (1998). 65. R. Alon, D. A. Hammer, T. A. Springer, Nature 374, 539(1995). 66. Y. C. Lee, R. T. Lee, Acc. Chem. Res. 28, 321 (1995). 67. R. Roy, Curr. Opin. Struct. Biol. 6, 692 (1996). 68. L. L. Kiessling, N. L. Pohl, Chem. Biol. 3, 71 (1996). 69. M. I. Page, W. P. Jencks, Proc. Natl. Acad. Sci. U.S.A. 68, 1678 (1971). 70. S. M. Dimick et al., J. Am. Chem. Soc. 121, 10286(1999). 71. S. D. Burke, Q. Zhao, M. C. Schuster, L. L. Kiessling,J. Am. Chem. Soc. 122, 4518 (2000). 72. E. K. Fan, E. A. Merritt, C. L. M. J. Verlinde, W. G. J. Hol,Curr. Opin. Struct. Biol. 10, 680 (2000). 73. J. Holmgren, A. M. Svennerholm, Gastroenterol. Clin.North Am. 21, 283 (1992). 74. E. K. Fan et al., J. Am. Chem. Soc. 122, 2663 (2000). 75. P. I. Kitov et al., Nature 403, 669 (2000). 76. W. F. Goebel, O. T. Avery, J. Exp. Med. 50, 521 (1929). 77. R. U. Lemieux, D. R. Bundle, D. A. Baker, J. Am. Chem.Soc. 97, 4076 (1975). 78. V. Pozsgay, Angew. Chem. Int. Ed. 37, 138 (1998). 79. S. J. Danishefsky, J. R. Allen, Angew. Chem. Int. Ed. 39,837 (2000). 80. E. J. Gordon, W. J. Sanders, L. L. Kiessling, Nature 392,30 (1998). 81. E. J. Gordon, L. E. Strong, L. L. Kiessling, Bioorg. Med.Chem. Lett. 6, 1293 (1998). 82. S. D. Rosen, Am. J. Pathol. 144, 1013 (1999). 83. T. K. Kishimoto, M. A. Jutila, E. L. Berg, E. C. Butcher,Science 245, 1238 (1989). 84. W. J. Sanders, E. J. Gordon, P. J. Beck, R. Alon, L. L.Kiessling, J. Biol. Chem. 274, 5271 (1999). 85. S. Goon, C. R. Bertozzi, ÒMetabolic substrate engineeringas a tool for glycobiology,Ó in Glycochemistry:Principles, Synthesis and Applications, P. G. Wang,C. R. Bertozzi, Eds. (Dekker, New York), in press. 86. A. K. Sarkar, T. A. Fritz, W. H. Taylor, J. D. Esko, Proc.Natl. Acad. Sci. U.S.A. 92, 3323 (1995). 87. F. N. Lugemwa, J. D. Esko, J. Biol. Chem. 266, 6674 (1991). 88. H. Kayser et al., J. Biol. Chem. 267, 16934 (1992). 89. O. T. Keppler et al., J. Biol. Chem. 270, 1308 (1995). 90. O. T. Keppler et al., Biochem. Biophys. Res. Commun.253, 437 (1998). 91. J. R. Wieser, A. Heisner, P. Stehling, F. Oesch, W.Reutter, FEBS Lett. 395, 170 (1996). 92. B. E. Collins, T. J. Fralich, S. Itonori, Y. Ichikawa, R. L.Schnaar, Glycobiology 10, 11 (2000). 93. L. K. Mahal, K. J. Yarema, C. R. Bertozzi, Science 276,1125 (1997). 94. E. Saxon, C. R. Bertozzi, Science 287, 2007 (2000). 95. K. J. Yarema, L. K. Mahal, R. Bruehl, E. C. Rodriguez,C. R. Bertozzi, J. Biol. Chem. 273, 31168 (1998). 96. G. A. Lemieux, K. J. Yarema, C. L. Jacobs, C. R. Bertozzi,J. Am. Chem. Soc. 121, 4278 (1999). 97. J. H. Lee et al., J. Biol. Chem. 274, 21878 (1999). 98. H. C. Hang, C. R. Bertozzi, J. Am. Chem. Soc. 123,1242 (2001). 99. The authors thank J. Gestwicki, L. Strong, E. Saxon, andK. Yarema for their comments on the manuscript, andthe NIH and NSF for research support.