Chapter 2: Simple Linear Regression • 2.1 The Model • 2.2
advertisement

Chapter 2: Simple Linear Regression
•
•
•
•
•
•
•
•
2.1 The Model
2.2-2.3 Parameter Estimation
2.4 Properties of Estimators
2.5 Inference
2.6 Prediction
2.7 Analysis of Variance
2.8 Regression Through the Origin
2.9 Related Models
1
2.1 The Model
• Measurement of y (response) changes in a linear fashion with a
setting of the variable x (predictor):
β0 + β1x
ε
y=
+
linear relation
noise
◦ The linear relation is deterministic (non-random).
◦ The noise or error is random.
• Noise accounts for the variability of the observations about the
straight line.
No noise ⇒ relation is deterministic.
Increased noise ⇒ increased variability.
2
• Experiment with this simulation program:
simple.sim <function(intercept=0,slope=1,x=seq(1,10),sigma=1){
noise <- rnorm(length(x),sd = sigma)
y <- intercept + slope*x + noise
title1 <- paste("sigma = ", sigma)
plot(x,y,pch=16,main=title1)
abline(intercept,slope,col=4,lwd=2)
}
• >
>
>
>
>
source("simple.sim.R")
simple.sim(sigma=.01)
simple.sim(sigma=.1)
simple.sim(sigma=1)
simple.sim(sigma=10)
3
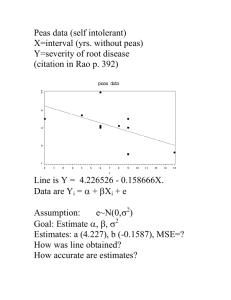
Simulation Examples
sigma = 0.1
6
4
y
6
2
2
4
y
8
8
10
10
sigma = 0.01
4
6
8
10
2
4
6
8
x
x
sigma = 1
sigma = 10
10
10 20 30
y
6
0
4
2
y
8
10
2
2
4
6
x
8
10
2
4
6
8
10
x
4
The Setup
• Assumptions:
1. E[y|x] = β0 + β1x.
2. Var(y|x) = Var(β0 + β1x + ε|x) = σ 2.
• Data: Suppose data y 1, y 2, . . . , y n are obtained at settings
x1, x2, . . . , xn, respectively. Then the model on the data is
y i = β0 + β1xi + εi
(εi i.i.d. N(0, σ 2) and E[y i|xi] = β0 + β1xi.)
Either
1. the x’s are fixed values and measured without error
(controlled experiment)
OR
2. the analysis is conditional on the observed values of x
(observational study).
5
2.2-2.3 Parameter Estimation, Fitted Values and Residuals
1. Maximum Likelihood Estimation
Distributional assumptions are required
2. Least Squares Estimation
Distributional assumptions are not required
6
2.2.1 Maximum Likelihood Estimation
◦ Normal assumption is required:
−1 (y −β −β x )2
1
f (yi|xi) = √
e 2σ2 i 0 1 i
2πσ
Likelihood: L(β0, β1, σ) =
1 − 12
∝ n e 2σ
σ
Y
f (yi|xi)
Pn
2
i=1 (yi −β0 −β1 xi )
◦ Maximize with respect to β0, β1, and σ 2.
◦ (β0, β1): Equivalent to minimizing
n
X
(yi − β0 − β1xi)2
i=1
(i.e. Least-squares) σ 2: SSE/n, biased.
7
2.2.2 Least Squares Estimation
◦ Assumptions:
1. E[εi] = 0
2. Var(εi) = σ 2
3. εi’s are independent.
• Note that normality is not required.
8
Method
• Minimize
S(β0, β1) =
n
X
(y i − β0 − β1xi)2
i=1
with respect to the parameters or regression coefficients β0 and
β1: βb0 and βb1.
◦ Justification: We want the fitted line to pass as close to all of the
points as possible.
◦ Aim: small Residuals (observed - fitted response values):
ei = y i − βb0 − βb1xi
9
Look at the following plots:
>
>
>
>
>
source("roller2.plot")
roller2.plot(a=14,b=0)
roller2.plot(a=2,b=2)
roller2.plot(a=12, b=1)
roller2.plot(a=-2,b=2.67)
10
−ve residual
4
6
8
60
40
0
2
4
6
8
10 12
a=12, b=1
a=−2, b=2.67
4
6
8
10 12
Roller weight (t)
60
40
20
20
0
−ve residual
Fitted values
Data values
+ve residual
−ve residual
0
40
60
−ve residual
Roller weight (t)
+ve residual
2
+ve residual
Roller weight (t)
Fitted values
Data values
0
20
10 12
Depression in lawn (mm)
2
Fitted values
Data values
0
+ve residual
Depression in lawn (mm)
20
40
60
Fitted values
Data values
0
Depression in lawn (mm)
a=2, b=2
0
Depression in lawn (mm)
a=14, b=0
0
2
4
6
8
10 12
Roller weight (t)
◦ The first three lines above do not pass as close to the plotted
points as the fourth, even though the sum of the residuals is
about the same in all four cases.
◦ Negative residuals cancel out positive residuals.
The Key: minimize squared residuals source("roller3.plot.R")
> roller3.plot(14,0); roller3.plot(2,2); roller3.plot(12,1);
> roller3.plot(a=-2,b=2.67) # small SS
4
6
8
120
80
40
2
4
6
8
10 12
0
a=12, b=1
a=−2, b=2.67
4
6
8
10 12
Roller weight (t)
80
120
Fitted values
Data values
40
40
80
120
0
Roller weight (t)
+ve residual
0
+ve residual
−ve residual
Roller weight (t)
−ve residual
2
Depression in lawn (mm)
10 12
Fitted values
Data values
0
Fitted values
Data values
+ve residual
−ve residual
0
2
Depression in lawn (mm)
120
80
40
+ve residual
−ve residual
0
Depression in lawn (mm)
a=2, b=2
Fitted values
Data values
0
Depression in lawn (mm)
a=14, b=0
0
2
4
6
8
10 12
Roller weight (t)
11
Unbiased estimate of σ 2
n
X
1
e2
n − # parameters estimated i=1 i
n
1 X
e2
=
n − 2 i=1 i
* n observations ⇒ n degrees of freedom
* 2 degrees of freedom are required to estimate the parameters
* the residuals retain n − 2 degrees of freedom
12
Alternative Viewpoint
* y = (y 1, y 2, . . . , y n) is a vector in n-dimensional space.
(n degrees of freedom)
* The fitted values ybi = βb0 + βb1xi also form a vector in
n-dimensional space:
b = (yb1, yb2, . . . , ybn)
y
(2 degrees of freedom)
b.
* Least-squares seeks to minimize the distance between y and y
* The distance between n-dimensional vectors u and v is the
square root of
n
X
(ui − vi)2
i=1
b is
* Thus, the squared distance between y and y
n
X
i=1
(y i − ybi)2 =
n
X
e2
i
i=1
13
Regression Coefficient Estimators
• The minimizers of
S(β0, β1) =
n
X
(y i − β0 − β1xi)2
i=1
are
βb0 = ȳ − βb1x̄
and
Sxy
b
β1 =
Sxx
where
Sxy =
n
X
(xi − x̄)(y i − ȳ)
i=1
and
Sxx =
n
X
(xi − x̄)2
i=1
14
HomeMade R Estimators
> ls.est <function (data)
{
x <- data[,1]
y <- data[,2]
xbar <- mean(x)
ybar <- mean(y)
Sxy <- S(x,y,xbar,ybar)
Sxx <- S(x,x,xbar,xbar)
b <- Sxy/Sxx
a <- ybar - xbar*b
list(a = a, b = b, data=data)
}
> S <function (x,y,xbar,ybar)
{
sum((x-xbar)*(y-ybar))
}
15
Calculator Formulas and Other Properties
•
Sxy =
n
X
Pn
Pn
( i=1 xi)( i=1 y i)
xiy i −
n
i=1
n
X
Pn
(
xi ) 2
2
i=1
Sxx =
xi −
n
i=1
• Linearity Property:
Sxy =
n
X
y i(xi − x̄)
i=1
so βb1 is linear in the y i’s.
• Gauss-Markov Theorem: Among linear unbiased estimators for
β1 and β0, βb1 and βb0 are best (i.e. they have smallest variance).
. Is it
• Exercise: Find the expected value and variance of xy1−ȳ
1 −x̄
unbiased for β1? Is there a linear unbiased estimator with
smaller variance?
16
Residuals
εbi = ei = y i − βb0 − βb1xi
= y i − ybi
= y i − ȳ − βb1(xi − x̄)
• In R:
> res
function (ls.object)
{
a <- ls.object$a
b <- ls.object$b
x <- ls.object$data[,1]
y <- ls.object$data[,2]
resids <- y - a - b*x
resids
}
17
2.3.1 Consequences of Least-Squares
1. ei = y i − ȳ − βb1(xi − x̄)
(follows from intercept formula)
P
2. n
ei = 0
(follows from 1.)
i=1
P
Pn
bi
3. n
y
=
i=1 i
i=1 y
(follows from 2.)
4. The regression line passes through the centroid (x̄, ȳ)
from intercept formula)
P
5.
xiei = 0
(set partial derivative of S(β0, β1) wrt β1 to 0)
P
6.
ybiei = 0
(follows from 2. and 5.)
(follows
18
2.3.2 Estimation of σ 2
• The residual sum of squares or error sum of squares is given by
SSE =
n
X
b
e2
i = Syy − β1 Sxy
i=1
Note: SSE = SSRes and Syy = SST
• An unbiased estimator for the error variance is
b 2 = M SE =
σ
SSE
=
n−2
h
Syy − βb1Sxy
i
n−2
Note: M SE = M SRes
b = Residual Standard Error =
• σ
√
MSE
19
Example
• roller data
weight depression
1
1.9
2
2
3.1
1
3
3.3
5
4
4.8
5
5
5.3
20
6
6.1
20
7
6.4
23
8
7.6
10
9
9.8
30
10
12.4
25
20
Hand Calculation
• (y = depression, x = weight)
P10
◦ i=1 xi = 1.9 + 3.1 + · · · 12.4 = 60.7
◦ x̄ = 60.7
10 = 6.07
P10
◦ i=1 y i = 2 + 1 + · · · 25 = 141
◦ ȳ = 141
10 = 14.1
◦
P10
2 = 1.92 + 3.12 + · · · + 12.42 = 461
x
i=1 i
P10
◦ i=1 y 2
i = 4 + 1 + 25 + · · · + 625 = 3009
◦
P10
i=1 xi y i = (1.9)(2) + · · · (12.4)(25) = 1103
(60.7)2
◦ Sxx = 461 − 10 = 92.6
21
◦ Sxy = 1103 − (60.7)(141)
= 247
10
(141)2
◦ Syy = 3009 − 10 = 1021
Sxy
247 = 2.67
= 92.6
◦ βb1 = Sxx
◦ βb0 = ȳ − β1x̄ = 14.1 − 2.67(6.07) = −2.11
b 2 = 1 (Syy − βb1Sxy )
◦ σ
n−2
=1
8 (1021 − 2.67(247)) = 45.2 = MSE
• Example Summary: the fitted regression line relating depression
(y) to weight (x) is
yb = −2.11 + 2.67x
The error variance is estimated as MSE = 45.2.
R commands (home-made version)
> roller.obj <- ls.est(roller)
> roller.obj[-3]
# intercept and slope estimate
$a
[1] -2.09
$b
[1] 2.67
> res(roller.obj)
# residuals
[1] -0.98 -5.18 -1.71 -5.71 7.95
[6] 5.82 8.02 -8.18 5.95 -5.98
> sum(res(roller.obj)ˆ2)/8 # error variance (MSE)
[1] 45.4
22
R commands (built-in version)
> attach(roller)
> roller.lm <- lm(depression ˜ weight)
> summary(roller.lm)
Call:
lm(formula = depression ˜ weight)
Residuals:
Min
1Q Median
-8.180 -5.580 -1.346
3Q
5.920
Max
8.020
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -2.0871
4.7543 -0.439 0.67227
weight
2.6667
0.7002
3.808 0.00518
23
Residual standard error: 6.735 on 8 degrees of freedom
Multiple R-squared: 0.6445,
Adjusted R-squared: 0.6001
F-statistic: 14.5 on 1 and 8 DF, p-value: 0.005175
> detach(roller)
• or
> roller.lm <- lm(depression ˜ weight, data = roller)
> summary(roller.lm)
Using Extractor Functions
• Partial Output:
> coef(roller.lm)
(Intercept)
weight
-2.09
2.67
> summary(roller.lm)$sigma
[1] 6.74
• From the output,
◦ the slope estimate is βb1 = 2.67.
◦ the intercept estimate is βb0 = −2.09.
◦ the Residual standard error is the square root of the MSE:
6.742 = 45.4.
24
Other R commands
◦ fitted values: predict(roller.lm)
◦ residuals: resid(roller.lm)
◦ diagnostic plots: plot(roller.lm)
(these include a plot of the residuals against the fitted values
and a normal probability plot of the residuals)
◦ Also plot(roller); abline(roller.lm)
(this gives a plot of the data with the fitted line overlaid)
25
2.4: Properties of Least Squares Estimates
◦ E[βb1] = β1
◦ E[βb0] = β0
2
σ
b
◦ Var(β1) = Sxx
x̄2
1+
◦ Var(βb0) = σ 2 n
Sxx
26
Standard Error Estimators
d βb ) = MSE
◦ Var(
1
Sxx
so the standard error (s.e.) of βb1 is estimated by
s
MSE
Sxx
roller e.g.: MSE
q = 45.2, Sxx = 92.6
⇒ s.e. of βb1 is 45.2/92.6 = .699
x̄2
d βb ) = MSE 1 +
◦ Var(
0
n
Sxx
so the standard error (s.e.) of βb0 is estimated by
v
u
u
tMSE
x̄2
1
+
n
Sxx
!
roller e.g.:
MSE = 45.2, Sxx
s = 92.6, x̄ = 6.07, n = 10
⇒ s.e. of βb0 is
1 +
45.2 10
6.072
92.62
= 4.74
27
Distributions of βb1 and βb0
◦ y i is N (β0 + β1xi, σ 2), and
P
xi −x̄
βb1 = n
a
y
(a
=
i
i
i
i=1
Sxx )
2
σ
b
⇒ β1 is N (β1, Sxx ).
2
b ) so
Also, SSE
is
χ
(independent
of
β
1
2
n−2
σ
βb1 − β1
∼ tn−2
q
M SE/Sxx
Pn
1 − x̄(xi −x̄) )
b
◦ β0 = i=1 ciyi
(ci = n
Sxx
1 + x̄2 ) and
⇒ βb0 is N (β0, σ 2 n
Sxx
βb0 − β0
s
MSE
1 + x̄2
n
Sxx
∼ tn−2
28
2.5 Inferences about the Regression Parameters
• Tests
• Confidence Intervals
for β1, and β0 + β1x0
29
2.5.1 Inference for β1
H0 : β1 = β10 vs. H1 : β1 6= β10
Under H0,
βb1 − β10
t0 = q
M SE/Sxx
has a t-distribution on n − 2 degrees of freedom.
p-value = P (|tn−2| > t0)
e.g. testing significance of regression for roller:
H0 : β1 = 0 vs. H1 : β1 6= 0
t0 =
2.67 − 0
= 3.82
.699
p-value = P (|t8| > 3.82) = 2(1 − P (t8 < 3.82)) = .00509
R command: 2*(1-pt(3.82,8))
30
(1 − α) Confidence Intervals
◦ slope:
βb1 ± tn−2,α/2s.e.
or
q
βb1 ± tn−2,α/2 MSE/Sxx
roller e.g. (95% confidence interval)
2.67 ± t8,.025(.699)
or
2.67 ± 2.31(.699) = 2.67 ± 1.61
R command: qt(.975, 8)
◦ intercept:
βb0 ± tn−2,α/2s.e.
e.g.
− 2.11 ± 2.31(4.74) = −2.11 ± 10.9
31
2.5.2 Confidence Interval for Mean Response
◦ E[y|x0] = β0 + β1x0
where x0 is a possible value that the predictor could take.
\ ] = βb + βb x is a point estimate of the mean response at
◦ E[y|x
0
0
1 0
that value.
◦ To find a 1 − α confidence interval for E[y|x0], we need the
\ ]:
variance of yb0 = E[y|x
0
Var(yb0) = Var(βb0 + βb1x0)
βb0 =
n
X
ciyi
i=1
βb1x0 =
n
X
bix0yi
i=1
32
Therefore,
n
X
yb0 =
(ci + bix0)yi
i=1
so
Var(yb0) =
n
X
(ci + bix0)2Var(yi)
i=1
= σ2
n
X
(ci + bix0)2
i=1
n
X
(x0 − x̄)(xi − x̄) 2
1
2
)
( +
=σ
Sxx
i=1 n
1
(x0 − x̄)2
2
=σ ( +
)
n
Sxx
1 + (x0 −x̄)2 )
\
◦ Var(
yb0) = MSE( n
Sxx
◦ The confidence interval is then
v
u
u
t
1
(x0 − x̄)2
yb0 ± tn−2,α/2 MSE( +
)
n
Sxx
◦ e.g. Compute a 95% confidence interval for the expected
depression when the weight is 5 tonnes.
x0 = 5, yb0 = −2.11 + 2.67(5) = 11.2
n = 10, x̄ = 6.07, Sxx = 92.6, MSE = 45.2
Interval:
s
1
(5 − 6.07)2
11.2 ± t8,.025 45.2(
+
)
10
92.6
√
= 11.2 ± 2.31 5.08 = 11.2 ± 5.21
= (5.99, 16.41)
◦ R code:
> predict(roller.lm,
newdata=data.frame(weight<-5),
interval="confidence")
fit lwr upr
[1,] 11.2 6.04 16.5
◦ Ex. Write your own R function to compute this interval.
2.6: Predicted Responses
◦ If a new observation were to be taken at x0, we would predict it
to be y 0 = β0 + β1x0 + ε where ε is independent noise.
◦ yb0 = βb0 + βb1x0 is a point prediction of the response at that
value.
◦ To find a 1 − α prediction interval for y 0, we need the variance of
y0 − yb0:
Var(y0 − yb0) = Var(β0 + β1x0 + ε − βb0 − βb1x0)
= Var(−βb0 − βb1x0) + Var(ε)
1
(x0 − x̄)2
2
=σ ( +
) + σ2
n
Sxx
33
1
(x0 − x̄)2
2
= σ (1 + +
)
n
Sxx
(x0 −x̄)2
1
\
◦ Var(y0 − yb0) = MSE(1 + n + Sxx )
◦ The prediction interval is then
v
u
u
t
(x0 − x̄)2
1
yb0 ± tn−2,α/2 MSE(1 + +
)
n
Sxx
◦ e.g. Compute a 95% prediction interval for the depression for a
single new observation where the weight is 5 tonnes.
x0 = 5, yb0 = −2.11 + 2.67(5) = 11.2
n = 10, x̄ = 6.07, Sxx = 92.6, MSE = 45.2
Interval:
s
1
(5 − 6.07)2
11.2 ± t8,.025 45.2(1 +
+
)
10
92.6
√
= 11.2 ± 2.31 50.3 = 11.2 ± 16.4 = (−5.2, 27.6)
R code for Prediction Intervals
> predict(roller.lm,
newdata=data.frame(weight<-5),
interval="prediction")
fit
lwr upr
[1,] 11.2 -5.13 27.6
◦ Write your own R function to produce this interval.
34
Degrees of Freedom
• A random sample of size n coming from a normal population
with mean µ and variance σ 2 has n degrees of freedom:
Y1, . . . , Yn.
• Each linearly independent restriction reduces the number of
degrees of freedom by 1.
Pn
(Yi −µ)2
• i=1 σ2
distribution.
has a χ2
(n)
Pn
(Yi −Ȳ )2
• i=1 σ2
has a χ2
distribution. (Calculating Ȳ imposes
(n−1)
one linear restriction.)
Pn
(Yi −Ybi )2
2
b and βb
• i=1
has
a
χ
distribution.
(Calculating
β
0
1
2
(n−2)
σ
imposes two linearly independent restrictions.)
• Given the xi’s, there are 2 degrees of freedom in the quantity
βb0 + βb1xi.
• Given xi and Ȳ , there is one degree of freedom:
ybi − Ȳ = βb1(xi − x̄).
35
2.7: Analysis of Variance: Breaking down (analyzing) Variation
◦ The variation in the data (responses) is summarized by
Syy =
n
X
(y i − ȳ)2 = TSS
i=1
(Total sum of squares)
◦ 2 sources of variation in the responses:
1. variation due to the straight line relationship with the
predictor
2. deviation from the line (noise)
◦
y i −ȳ
deviation from data center
y i −ybi
i −ȳ
= residual
+ difference:ybline
and center
y i − ȳ = ei + ybi − ȳ
36
so
Syy =
=
=
(y i − ȳ)2
X
(ei + ybi − ȳ)2
X
X
e2
i +
(ybi − ȳ)2
X
since
X
eiybi = 0 and ȳ
Syy = SSE +
X
ei = 0
(ybi − ȳ)2 = SSE + SSR
X
The last term is the regression sum of squares.
Relation between SSR and β1
◦ We saw earlier that
SSE = Syy − βb1Sxy
◦ Therefore,
SSR = βb1Sxy = βb12Sxx
Note that, for a given set of x’s SSR depends only on βb1.
M SR = SSR /d.f. = SSR /1
(1 degree of freedom for slope parameter)
37
Expected Sums of Squares
◦
E[SSR ] = E[Sxxβb12]
= Sxx Var(βb1) + (E[βb1])2
= Sxx
σ2
!
+ β12
Sxx
= σ 2 + β12Sxx
= E[M SR ]
Therefore, if β1 = 0, then SSR is an unbiased estimator for σ 2.
◦ Development of E[MSE]:
E[Syy ] = E[
= E[
X
(yi − ȳ)2]
X
yi2 − nȳ 2] =
X
E[yi2] − nE[ȳ 2]
38
Development of E[MSE] (cont’d)
Consider the 2 terms on RHS, separately:
1. term:
E[yi2] = Var(yi) + (E[yi])2
= σ 2 + (β0 + β1xi)2
X
E[yi2] = nσ 2 + nβ02 + 2nβ0β1x̄ +
X
β12x2
i
2. term:
E[ȳ 2] = Var(ȳ) + (E[ȳ])2
= σ 2/n + (β0 + β1x̄)2
nE[ȳ 2] = σ 2 + nβ02 + 2nβ0β1x̄ + nβ12x̄2
39
Development of E[MSE] (cont’d)
⇒
E[Syy ] = (n − 1)σ 2 + β12
(xi − x̄)2
X
= E[SST ]
◦
E[SSE] = E[Syy ] − E[SSR ]
= (n − 1)σ 2 + β12
(xi − x̄)2 − (σ 2 + β12Sxx)
X
= (n − 2)σ 2
E[MSE] = E[SSE/(n − 2)] = σ 2
40
Another approach to testing H0 : β1 = 0
◦ Under the null hypothesis, both MSE and M SR estimate σ 2.
◦ Under the alternative, only MSE estimates σ 2.
E[M SR ] = σ 2 + β12Sxx > σ 2
◦ A reasonable test is
F0 =
M SR
MSE
∼ F1,n−2
◦ Large F0 ⇒ evidence against H0.
◦ Note t2
ν = F1,ν so this is really the same test as
t2
0 = q
βb1
MSE/Sxx
2
βb12Sxx
M SR
=
=
MSE
M SE
41
The ANOVA table
Source
Reg.
df
1
SS
βb12Sxx
Syy − βb12Sxx
Syy
MS
βb12Sxx
Error
n−2
SSE/(n − 2)
Total
n-1
roller data example:
> anova(roller.lm) # R code
Analysis of Variance Table
F
M SR
MSE
Response: depression
Df Sum Sq Mean Sq F value Pr(>F)
weight
1
658
658
14.5 0.0052
Residuals 8
363
45
(recall that the t-statistic for testing β1 = 0 had been 3.81 =
√
14.5)
◦ Ex. Write an R function to compute these ANOVA quantities.
42
Confidence Interval for σ 2
2
∼
χ
◦ SSE
2
n−2
σ
so
SSE
2
2
P (χn−2,1−α/2 ≤
≤
χ
)=1−α
n−2,α/2
2
σ
so
SSE
P( 2
χn−2,α/2
≤ σ2 ≤
SSE
χ2
n−2,1−α/2
)=1−α
e.g. roller data:
SSE = 363
χ2
8,.975 = 2.18
(R code: 1-qchisq(8, .025))
χ2
8,.025 = 17.5
(363/17.5, 363/2.18) = (20.7, 166.5)
43
2.7.1 R2 - Coefficient of Determination
◦ R2 = is the fraction of the response variability explained by the
regression:
SSR
2
R =
Syy
◦ 0 ≤ R2 ≤ 1. Values near 1 imply that most of the variability is
explained by the regression.
◦ roller data:
SSR = 658 and Syy = 1021
so
658
2
R =
= .644
1021
44
R output
> summary(roller.lm)
...
Multiple R-Squared: 0.644, ...
◦ Ex. Write an R function which computes R2.
◦ Another interpretation:
β12Sxx + σ 2
. E[SSR ]
2
=
E[R ] =
E[Syy ]
(n − 1)σ 2 + β12Sxx
2
2 Sxx
Sxx + σ
β
β12 n−1
1 (n−1)
n−1 .
=
=
Sxx
Sxx
σ 2 + β12 n−1
σ 2 + β12 (n−1)
for large n. (Note: this differs from the textbook.)
45
Properties of R2
• Thus, R2 increases as
1. Sxx increases (x’s become more spread out)
2. σ 2 decreases
◦ Cautions
1. R2 does not measure the magnitude of the regression slope.
2. R2 does not measure the appropriateness of the linear model.
3. A large value of R2 does not imply that the regression model
will be an accurate predictor.
46
Hazards of Regression
• Extrapolation: predicting y values outside the range of observed
x values. There is no guarantee that a future response would
behave in the same linear manner outside the observed range.
e.g. Consider an experiment with a spring. The spring is
stretched to several different lengths x (in cm) and the restoring
force F (in Newtons) is measured:
x
F
3
5.1
4
6.2
5
7.9
6
9.5
> spring.lm <- lm(F˜x - 1,data=spring)
> summary(spring.lm)
Coefficients:
47
Estimate Std. Error t value Pr(>|t|)
x
1.5884
0.0232
68.6 6.8e-06
The fitted model relating F to x is
Fb = 1.58x
Can we predict the restoring force for the spring, if it has been
extended to a length of 15 cm?
• High leverage observations: x values at the extremes of the
range have more influence on the slope of the regression than
observations near the middle of the range.
• Outliers can distort the regression line. Outliers may be
incorrectly recorded OR may be an indication that the linear
relation or constant variance assumption is incorrect.
• A regression relationship does not mean that there is a
cause-and-effect relationship. e.g. The following data give the
number of lawyers and number of homicides in a given year for a
number of towns:
no. lawyers
1
2
7
10
12
14
15
18
no. homicides
0
0
2
5
6
6
7
8
Note that the number of homicides increases with the number of
lawyers. Does this mean that in order to reduce the number of
homicides, one should reduce the number of lawyers?
• Beware of nonsense relationships. e.g. It is possible to show
that the area of some lakes in Manitoba is related to elevation.
Do you think there is a real reason for this? Or is the apparent
relation just a result of chance?
2.9 - Regression through the Origin
◦ intercept = 0
y i = β1xi + ε
◦ Max. Likelihood and L-S ⇒
minimize
n
X
(yi − β1xi)2
i=1
P
xy
βb1 = P i2i
xi
ei = y i − βb1xi
SSE =
X
e2
i
◦ Max. Likelihood ⇒
b2 =
σ
SSE
n
48
◦ Unbiased Estimator:
b 2 = MSE =
σ
SSE
n−1
◦ Properties of βb1:
E[βb1] = β1
2
σ
Var(βb1) = P 2
xi
◦ 1 − α C.I. for β1:
v
u
u MSE
b
β1 ± tn−1,α/2t P 2
xi
◦ 1 − α C.I. for E[y|x0]:
v
u
u MSEx2
yb0 ± tn−1,α/2t P 20
xi
since
2
σ
Var(βb1x0) = P 2 x2
xi 0
◦ 1 − α P.I. for y, given x0:
v
u
u
x2
t
yb0 ± tn−1,α/2 MSE(1 + P 02 )
xi
◦ R code:
> roller.lm <- lm(depression˜weight - 1,
data=roller)
> summary(roller.lm)
Coefficients:
Estimate Std. Error t value Pr(>|t|)
weight
2.392
0.299
7.99 2.2e-05
Residual standard error: 6.43
on 9 degrees of freedom
Multiple R-Squared: 0.876,
Adjusted R-squared: 0.863
F-statistic: 63.9 on 1 and 9 DF,
p-value: 2.23e-005
> predict(roller.lm,
newdata=data.frame(weight<-5),
interval="prediction")
fit
lwr upr
[1,] 12.0 -2.97 26.9
Ch. 2.9.2 Correlation
• Bivariate Normal Distribution:
f (x, y) =
1
−
q
2πσ1σ2 1 − ρ2
e
1
(Y 2 −2ρXY +X 2 )
2
2(1−ρ )
where
x − µ1
X=
σ1
and
Y =
y − µ2
σ2
◦ correlation coefficient:
σ12
ρ=
σ1σ2
◦ µ1 and σ12 are the mean and variance of x
◦ µ2 and σ22 are the mean and variance of y
◦ σ12 = E[(x − µ1)(y − µ2)], the covariance
49
Conditional distribution of y given x
f (y|x) = √
1
2πσ1.2
y−β0 −β1 x 2
1
−
σ1.2
e 2
where
σ
β0 = µ2 − µ1ρ 2
σ1
σ
β1 = 2 ρ
σ1
2 = σ 2 (1 − ρ2 )
σ1.2
2
50
An Explanation
◦ Suppose
:
y = β0 + β1x + ε
where ε and x are independent N (0, σ 2) and N (µ1, σ12) random
variables.
◦ Define µ2 = E[y] and σ22 = Var(y):
µ2 = β0 + β1µ1
σ22 = β12σ12 + σ 2
◦ Define σ12 = Cov(x − µ1, y − µ2):
σ12 = E[(x − µ1)(y − µ2)]
= E[(x − µ1)(β1x + ε − β1µ1)]
= β1E[(x − µ1)2] + E[(x − µ1)ε]
= β1σ12
51
◦ Define ρ = σσ12
:
1 σ2
β1σ12
σ
= β1 1
ρ=
σ1σ2
σ2
Therefore,
σ2
ρ
β1 =
σ1
and
β0 = µ2 − β1µ1
σ
= µ2 − µ1 2 ρ
σ1
◦ What is the conditional distribution of y, given x = x?
y|x=x = β0 + β1x + ε
must be normal with mean
E[y|x = x] = β0 + β1x
σ2
= µ2 + ρ (x − µ1)
σ1
and variance
2 = σ 2 = σ 2 − β 2σ 2
Var(y|x = x) = σ1.2
2
1 1
2
σ
= σ22 − ρ2 22 σ12
σ1
= σ22(1 − ρ2)
Estimation
◦ Maximum Likelihood Estimation (using bivariate normal) gives
βb0 = ȳ − βb1x̄
βb1 =
Sxy
Sxx
r = ρb = q
Sxy
SxxSyy
◦ Note that
s
r = βb1
Sxx
Syy
so
Sxx
2
2
b
r = β1
= R2
Syy
52
i.e. the coefficient of determination = square of correlation
coefficient
Testing H0 : ρ = 0 vs. H1 : ρ 6= 0
◦ Cond’l approach (equiv. to testing β1 = 0):
βb1
t0 = q
M SE/Sxx
=r
βb1
Syy −βb1 2 Sxx
(n−2)Sxx
v
u 2
u βb (n − 2)
1
=u
t S
yy
b 2
−
β
1
Sxx
v
u
u (n − 2)r 2
=t
1 − r2
where we have used βb12 = r2Syy /Sxx.
53
◦ The above statistic is a t statistic on n − 2 degrees of freedom ⇒
conclude ρ 6= 0 if pvalue
P (|tn−2| > |t0|) is small.
Testing H0 : ρ = ρ0
1+r
1
log
2
1−r
has an approximate normal distribution with mean
Z=
µZ =
1
1+ρ
log
2
1−ρ
and variance
2 =
σZ
1
n−3
for large n.
◦ Thus,
Z0 =
1+ρ0
log 1+r
−
log
1−r
1−ρ
0
q
2 1/(n − 3)
has an approximate standard normal distribution when the null
hypothesis is true.
54
Confidence Interval for ρ
1+ρ
• Confidence interval for 1
log
2
1−ρ :
q
Z ± zα/2 1/(n − 3)
• Find endpoints (l, u) of this confidence interval and solve for ρ:
e2l − 1 e2u − 1
(
,
)
2l
2u
1+e 1+e
55
R code for fossum example
• Find 95% confidence interval for the correlation between total
length and head length:
>
>
>
>
>
source("fossum.R")
attach(fossum)
n <- length(totlngth)
# sample size
r <- cor(totlngth,hdlngth)# correlation est.
zci <- .5*log((1+r)/(1-r)) +
qnorm(c(.025,.975))*sqrt(1/(n-3))
> ci <- (exp(2*zci)-1)/(1+exp(2*zci))
> ci
# transformed conf. interval
> detach(fossum)
[1] 0.62
0.83
# conf. interval for true
# correlation
56