will
advertisement

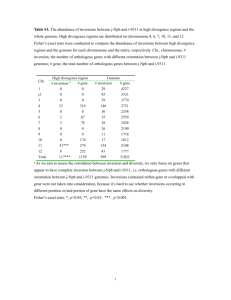
Pericentric Inversions (around the centromere) Kinds of chromatids produced by various crossovers within the inversion in a pericentric inversion heterozygote C Constitution i i off 4 chromatids h id after f C.O. CO C.O. event Position of C.O. Normal Single C.O CO Inversion dp + df Any point 1 1 2 2 Strand 2-Strand 12 1,2 2 2 0 3-Strand 1,3 or 1,4 1 1 2 4 St d 4-Strand 15 1,5 0 0 4 Double The numbers in column 2 refer to those in inversion figure. g C.O.=Crossover Pericentric Inversions ( (around d the th centromere) t ) The two types of inversions (para- and peri-centric) result in different cytological events Chromosome inversions have no effect on mitotic divisions, but do effect meiosis Meiosis is normal in individuals with homozygous inversions If the inverted regions of the inversion heterozygote is large enough for crossing-over i to occur within i hi the h inversion i i loop, l a portion i off the h resulting li gametes will be abnormal Pericentric Inversions (around the centromere) Pericentric inversions result in dp + df gametes which is related to the chromosome segments distal to the breakpoints of the inverted segment Products of crossover event within the loop are lost 1. 2. 3. Recombinant types are not recovered Crossing over may not be suppressed cytologically Only 2 strand double crossover types are recovered No bridges or fragments produced at anaphase I or II Paracentric Inversions (beside the centromere) In plants p a ts A homozygous inversion will produce normal pollen and seed set A heterozygous inversion will produce partial ovule and pollen abortion The degree Th d off pollen ll abortion b ti is i dependent d d t upon the th amountt off crossing-over i within the inversion loop To distinguish a homozygous inversion from a homozygous normal individual, the unknown can be crossed with a homozygous normal individual If the unknown is a homozygous inversion, the F1 will be h t heterozygous for f the th inversion i i andd be b partially ti ll sterile t il Paracentric Inversions (beside the centromere) Crossing-over in paracentric inversions result in bridges (dicentric chromosomes) h ) andd fragments f t (acentric ( t i chromosomes) h ) The size of the acentric fragment represents the length of the inverted region plus twice the length of the distal segment If deficiencies for the segment distal to the inversion, resulting from the loss of the acentric fragment, cause gamete spore abortion, the pollen abortion percentage can be predicted by cytological observation of meiosis Paracentric Inversions (besides the centromere) Kinds of chromatids produced by various crossovers in a paracentric inversion heterozygote Constitution of 4 chromatids after C.O. C.O. event Position of C.O. Normal Single C.O Inversion dp + df Any point 1 1 2 (dicentric+acentric) 2-Strand 1,2 2 2 0 3-Strand 1,3 or 1,4 1 1 2 (dicentric+acentric) 4-Strand 1,5 0 0 4 (dicentric+acentric) IN/OUT 1 in loop + 1 in interstitial 1 1 2 (dicentric+acentric) IN/OUT 44-strand strand double + 1 in interstitial 0 0 4 (dicentric+acentric) Double Chromatid tie in Dorsophila female Oogenesis Division vso I A dicentric bridge orients crossover chromatids away from the poles at division I Deficiency-duplication chromatids will occur in intercalary cell and polar cells will produce a fertile ovum with intact chromosomes Division v s o II During female gamete formation in plants and animals, animals only one polar megaspore will function in production of the ovule The duplication-deficiency p y chromosomes are oriented to the intercalary cells by the chromatid tie and the normal chromosomes will be included in the nuclei of the polar cells more often than the dp-df chromosomes The result will be nearly normal female fertility of inversion heterozygotes, but recombination will be substantially reduced in regions i involved i l d in i the th inversion i i Use of inversions to produce duplications without deficiencies Intercross two inversions with one breakpoint in common: 1 2 3 4 5 6 7 8 1 3 2 4 5 6 7 8 1 X 2 1 4 3 4 5 6 7 8 3 2 5 6 7 8 4 1 3 2 5 6 7 8 1 3 2 5 6 7 8 1 4 3 2 4 1 3 2 5 6 7 8 4 5 6 7 8 Intercross overlapping inversions: 1 2 3 4 5 6 7 8 1 3 2 4 5 6 7 8 1 X 2 1 3 2 4 3 2 4 2 5 4 5 6 6 7 7 1 8 1 1 3 2 2 4 4 5 3 4 2 3 3 6 5 7 6 8 7 8 1 1 2 4 5 3 1 8 5 3 4 5 3 5 3 2 1 1 4 5 7 6 7 6 7 8 6 7 8 6 2 6 7 4 8 8 8 5 6 7 8 c b d e a A double crossover is required for recovery of chromatid with recombination within the inversion loop c c will have greatest amount of recombination with a a b and d will have equal amount of recombination with a, but less than c b d e Interchromosomal Translocation (part of one chromosome is attached to another) Types 1. Interstitial translocations (intercalary) A segment from one chromosome is transferred to a position in another chromosome. Requires three breaks. 2. Reciprocal translocation (interchange) Two non-homologous chromosomes have symmetrically exchanged segments. One break in each chromosome is sufficient. Nearly always involves terminal end segments. segments I t titi l segmentt = segmentt off an interchange Interstitial i t h chromosome h between b t the th breakpoint b k i t and the centromere. Interchromosomal Translocation A cross configuration is formed at pachytene of interchange heterozygotes. The position of the cross is a reflection of where the breakpoint has occurred. During diplotene and diakinesis, the chromosomes shorten, the chiasma terminalize, and the cross configuration opens up to form a ring of 4 if chiasma are present. a b 1 c d e f 2 g h a b 12 g h e f 21 c d d c 12 d b h a g 1 h b g a c Pachytene pairing of interchange heterozygote 21 e f 2 e f d d 1 c c 21 e f b a f a e 2 b g 12 g h h Interchromosomal Translocation Observed meiotic configurations depend on the occurrence of chiasmata No. of Arms with Chiasma Diakinesis Configuration 4 Ring of 4 (4) 3 Chain of 4 (IV, 4 types) 2 adjacent arms Chain of 3 + univalent (III+I, 4 types) 2 alternate arms 2 pairs (2II, (2II 2 types) Orientation of interchange heterozygote quadrivalent at Metaphase I d c 12 d b h a g 1 h b g a c Adjacent I 21 e f 2 e f Adjacent non non-homologous homologous centromeres pass to the same pole 1 + 21 12 + 2 dp cd + df gh dp gh + df cd Orientation of interchange heterozygote quadrivalent at Metaphase I Adjacent II a a b b 12 1 h g c d h g c d 21 2 e e Adjacent homologous centromeres pass to the same pole f f 1 +12 21+ 2 dp ab + df ef dp ef + df ab Orientation of interchange heterozygote quadrivalent at Metaphase I d c 12 d b h a g 1 h b g a c Alternate 21 e f 2 e f Alternate disjunction of non nonhomologous centromeres 1 +2 12 + 21 Normal Balanced translocation Disjunction from a ring quadrivalent Orientation of chromosomes of a ring of 4 may be either an open or a zig-zag zig zag configuration leading to either adjacent or alternate chromosome disjunction. Adjacent I disjunction Adjacent but non-homologous centromeres migrate to the same pole. 1+21 12+2 Dp p fe +Df jjk Dp jk +Df fe Dp=duplication p p Df=deficiency Gametes usually abort. Adjacent II disjunction Occurss rarely Occu a e y if eve ever.. Adjacent djace t but homologous o o ogous ce centromeres t o e es migrate g ate to the t e same sa e pole. po e. 1+12 21+2 Gametes abort. Dp abcd +Df ghi Dp ghi+Df abcd Disjunction from a ring quadrivalent Alternate disjunction Alternate centromeres migrate to the same pole at anaphase I. 1+ 2 12+ 21 Normal chromosome complement p Interchange chromosome complement Both combinations produce viable gametes. Factors influencing orientation of a ring quadrivalent Considering 2 normal bivalents, there is complete independence and adjacent I and alternate disjunction will occur with equal frequency. Adjacent II should be impossible since there is no opportunity for coorientation between non-homologous centromeres. With production of quadrivalent co-orientation co orientation of non-homologous non homologous centromeres becomes possible. With random co-orientation: alternate l disjunction di j i frequency f = adjacent dj disjunction di j i frequency f Even within species there is considerable genetic variation affecting the ratio of alternate and adjacent j disjunction. j In most cases either alternate or adjacent predominates so that co-orientation is not a reality. Random orientation may occur in early prophase but soon forces act on quadrivalent, changing the orientation of the quadrivalent. Factors influencing orientation of a ring quadrivalent Forces acting on the quadrivalent: 1. Contraction of chromosomes resulting in stiffness and torsion. Short stiff chromosomes or those with little tendency for chiasma terminalization do not have sufficient flexibility for alternate disjunction. 2. Centromere activity Centromere orientation is maintained by the presence of counter-force exerted on the centromere Alternate orientation p provided more stable counter forces and will not readily y revert to adjacent orientation. With adjacent orientation if the pull from a single opposite centromere lapses, both co-orienting centromeres become unstable and resume equal probabilities to orient to either pole. With time the alternate orientation often accumulates. In rye interchange heterozygotes, alternate rings may occur in up to 95% of PMCs in late metaphase. Factors influencing orientation of a ring quadrivalent 1. Forces acting on the quadrivalent. 2. Length of interchange and interstitial segment. 3. Localization and terminalization of chiasmata. Genetic consequence of interchange A iinterchange An t h behaves b h like lik a single i l genetic ti factor. f t Two reciprocal translocations that do not have a chromosome in common segregate independently. In the translocation homozygote, the linkage relationship will be changed. Genes in the translocated segment fail to show linkage with genes in the chromosome h where h they th originally i i ll occurred. d Products of Alternate disjunction a b 1 c d e f 2 g h a b 12 g h e f 21 c d Normal linkage map A Linkage g map p from pprogeny g y of translocation heterozygote E F B C G D H E F G A H C B D Id ifi i off chromosomes Identification h involved i l d in i interchanges i h 1. Cytology Pachytene y analysis y of chromosomes involved in the cross configuration g Karyotype analysis of somatic cells Unequal size of exchange segment allows identification of change in chromosome lengths Banding pattern of chromosomes Direct observation of Dorsophila salivary gland chromosome bands 2. Genetic Linkage Genes on one chromosome become linked to those on another Genes known to be linked or independent suddenly change relationship Identification of chromosomes involved in interchanges 1. Use of trisomic tester 2n 1 known trisomic tester lines are crossed with unknown interchange 2n+1 stocks If one of the chromosomes involved in the translocation is the trisomic chromosome, a chain of 5 is expected If th the ttrisomic i i does d nott involve i l one off the th interchange i t h chromosomes, h a ring of 4 plus a trivalent are expected 2 Chromosome identification set 2. Cross a series of known interchange stocks with the unknown interchange stock and examine the F1 at meiotic metaphase I Two rings of 4 indicate the interchanges are independent A ring of 6 indicates one chromosome of the interchange is in common with one of the tester interchange chromosomes An F1 from a cross between interchange g stocks involvingg the same two chromosomes will not produce an association larger than a ring of 4 or may produce mostly/only pairs