GE Healthcare
Application Note 28-9327-13 AA IN Cell Investigator
Analysis of F-actin network reorganization using
IN Cell Investigator
Key words: actin • cytoskeleton • organization •
phalloidin • Developer Toolbox • IN Cell Investigator
The cytoskeleton is a dynamic three-dimensional structure
contained within the cytoplasm of the cell. It provides
mechanical support, determines the shape of the cell, and
plays an important role in cell movement, replication and
differentiation. It also participates in intercellular junctions
and interactions between the cell and the extracellular
matrix. The cytoskeleton is comprised of an organized
network of three predominant protein families—namely
actin microfilaments, microtubules and intermediate
filaments. Abnormalities of these proteins and consequent
disruption of the cytoskeletal matrix are associated with
many diseases including cancer, neurodegeneration and
cardiovascular disease.
G-actin, the monomeric globular form of the actin protein, is
associated with ATP, and polymerizes in a helical manner
that utilizes ATP hydrolysis to form filaments or fibers of
F-actin (1). The filaments are polar, having faster
polymerizing barbed ends and slower growing pointed ends,
allowing filament elongation and treadmilling. In addition,
incorporation of actin binding proteins such as CapZ and
Arp2/3 leads to capping or crosslinking of growing filaments,
resulting in the formation of bundles of F-actin and a
network structure providing cellular shape and support (2).
Cell motility is achieved by the cooperation of protrusive and
contractile arrays of F-actin found in the lamellipodia and
filopodia of cells (3). All of these processes are controlled
primarily via reorganization of the actin cytoskeleton,
mediated by a variety of factors including Rac1, Cdc42, and
RhoA proteins. Activated Rac1 is involved in the formation of
lamellipodia and membrane ruffling, Cdc42 activation
generates filopodia, and activation of RhoA has been shown
to contribute to the formation of stress fibers (4).
This application note demonstrates the imaging and
analysis of F-actin fibers in three different cell types, using
the subcellular imaging capability of IN Cell Analyzer 1000
and the flexibility of IN Cell Investigator image analysis tools.
Disruption of actin polymerization or reorganization of the
actin cytoskeleton network due to specific compound
treatments are described, and results visualized using
Spotfire DecisionSite™.
Materials
Products used
IN Cell Analyzer 1000*
28-4051-28
IN Cell Investigator single seat license†
28-4089-71
IN Cell Investigator 1 additional seat license†
28-4089-75
IN Cell Investigator 5 seat license†
28-4089-72
*IN Cell Analyzer 3000 may also be used for these experiments.
†
A seat license is a cost-effective single-user or server license that gives
access to all ready-to-use Image Analysis Modules provided for your IN Cell
Analyzer instrument. License holders have access to all appropriate analysis
software and more licenses can be purchased as the number of users grows.
Other materials required
HeLa, NIH 3T3, and CHO cell lines‡
(ECACC)
Dulbecco’s Modified Eagle’s Medium supplemented (Sigma)
with 10% fetal bovine serum, 2 mM l-glutamine, and
100 µg/ml penicillin-streptomycin
Ham’s F-12 medium supplemented with 10%
(Sigma)
fetal bovine serum, 2 mM l-glutamine, and 100 µg/ml
penicillin-streptomycin
Cytochalasin D, staurosporine
(Sigma)
Hoechst™ 33342
(Molecular Probes)
Texas Red™-X phalloidin
(Molecular Probes)
µClear™ 96-well microplates, black (Greiner Bio-One GmbH)
Most countries have legislation governing the handling, use, storage,
disposal and transportation of mammalian cell lines. Readers must be aware
of and observe the Local Regulations or Codes of Practice, which relate to
such matters prior to experimentation.
‡
Methods
Assay
HeLa and NIH 3T3 cells were cultured in Dulbecco’s Modified
Eagle’s medium and CHO cells were cultured in Ham’s F12
medium. Cells in log-phase growth were seeded into 96-well
microplates (5000 cells/well) and incubated in culture
medium for 24 h at 37°C, 5% CO2. Culture medium was
removed from the cells and replaced with complete medium
(100 μl) containing test compounds. Following incubation
with the compounds for 2 h at 37°C, 5% CO2, cells were fixed
with formaldehyde (2%) for 20 min, permeabilized with 0.2%
Triton™ X-100 in PBS for 30 min, and then stained with
Hoechst 33342 (10 µM) and Texas Red-X phalloidin (1:100) in
PBS for 20 min.
Treatment of NIH 3T3 cells with the fungal metabolite
cytochalasin D induced disruption of the linear arrangement of
F-actin fibers, loss of fibers, and a perinuclear or speckled
accumulation of stained actin. Cells appeared to lose their
characteristic shapes and assume more rounded morphologies
as the F-actin network was disrupted (Fig 2B). In contrast,
staurosporine treatment of the same cell line resulted in
maintenance of the cell shape, whilst the linear structures (Fig 2A)
were replaced by a more diffuse pattern of F-actin staining in the
cytoplasm (Fig 2C). Stress fibers were lost and the majority of
F-actin staining was localized to the plasma membrane which
became wavy and almost ruffled in appearance compared to
the control cells. The maintenance of cell shape after
staurosporine treatment is reported to be due to the continued
presence of microtubules and intermediate filaments (5).
Imaging and analysis
Images were acquired on IN Cell Analyzer 1000 using a 20×
objective with 500 ms exposure times for both Hoechst
(360/40 nm excitation filter; 460/40 nm emission filter) and
Texas Red (570/20 nm excitation filter; 620/60 nm emission
filter). Images were analyzed using IN Cell Developer Toolbox
(part of IN Cell Investigator software). The fluorescent signal
from Hoechst 33342 was used as a marker to identify all
nuclei. Texas Red-conjugated phalloidin binds with higher
affinity to polymerized actin than monomeric actin, and is
therefore used to identify F-actin fibers. The analysis
protocol was designed to detect F-actin fibers present within
a defined sampling region of the cell, and to report a variety
of user-defined fiber-related measures.
Results
F-actin organization
HeLa, NIH 3T3, and CHO cells were stained with Texas
Red-labeled phalloidin to visualize the F-actin cytoskeleton
(Fig 1). Although there were subtle differences in the
observed cytoskeletal structure in the different cell types, all
exhibited an organized network of linear F-actin fibers
throughout the cytoplasm, forming a mostly parallel
arrangement. In CHO cells, F-actin staining was also more
pronounced at the cell boundaries.
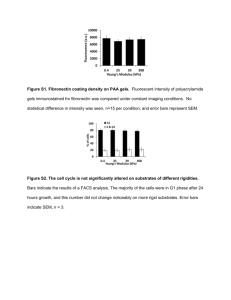
Fig 1. HeLa, NIH 3T3, and CHO cells were cultured in 96-well microplates
overnight before fixation and staining. Nuclei exhibit blue fluorescence due
to Hoechst 33342; F-actin exhibits red fluorescence due to Texas Redconjugated phalloidin.
2
28-9327-13 AA
Fig 2. Mouse Swiss NIH embryo cells (NIH 3T3) were treated with (A)
complete culture medium; (B) 10 µM cytochalasin D; or (C) 0.3 μM
staurosporine for 2 h, prior to fixation and staining. Nuclei exhibit blue
fluorescence due to Hoechst 33342 and F-actin exhibits red fluorescence
due to Texas Red-conjugated phalloidin.
Analysis of morphological changes
Treatment of the cell lines with cytochalasin D or
staurosporine resulted in a reduction in the presence of
visible fibers in the cytoplasm. This change was
quantitated by measuring the length of linear structures
using the fiber length per cell measure. The analysis
protocol was designed to detect the presence or
absence of these structures within a defined sampling
region. Marked differences between control and treated
cell populations were readily apparent when the data
were plotted (Fig 3).
Fig 3. HeLa (blue), CHO (green), and NIH 3T3 (red) cells were exposed to culture
medium (-) or 2.2 µM cytochalasin D (+) for 2 h prior to fixation and staining of
nuclei and F-actin. Bars represent the mean ±SD of n = 8 (HeLa, CHO) or n = 4 (NIH
3T3) replicate images. Magnitude of response (MOR) and signal:noise (S:N) data
are given for each cell type. Cell image shows Texas Red-conjugated phalloidin
staining of actin fibers in untreated NIH 3T3 cells, with colored overlays defining
actin fibers (green) and sampling region (cyan) as a result of analysis.
Dose response data
Data visualization
Treatment of HeLa, NIH 3T3 and CHO cells with increasing
concentrations of the agent cytochalasin D caused the organized
arrangement of F-actin fibers to become increasingly disrupted.
Dose response data show a reduction in actin fiber length for all
cell types with similar IC50 values for NIH 3T3 and CHO cells of
1.011 and 1.084 µM respectively and an IC50 value of 0.204 µM for
HeLa cells (Fig 4). These observations correlate with reports that
cytochalasin D binds reversibly to the growing end of the F-actin
filament, preventing further elongation (6). In addition,
cytochalasin D prevents the attachment of binding proteins,
thereby disrupting filament crosslinking and hence formation of
the F-actin fiber network (7).
Using the integrated links to Spotfire DecisionSite from
within Investigator software, it was possible to visualize the
data in a variety of ways (Fig 6). Dose response data for
cytochalasin D treatment were plotted and analyzed using
fiber and chord related measures. Data were also displayed
as a histogram of actin fiber length in relation to the
concentration of cytochalasin D. Shorter fibers of F-actin
(< 10 µm) were predominantly observed in wells
corresponding to higher concentrations of cytochalasin D
(> 500 nM, red bars). In contrast, longer actin fibers (> 30 µm)
were only present at low concentrations of cytochalasin D
(< 500 nM). Visualization of the data using the selected
measures enabled refinement of the analysis parameters
and also assisted in data interpretation.
Fig 4. Dose response curves for inhibition of F-actin fiber formation with
cytochalasin D treatment. HeLa (green), NIH 3T3 (blue) and CHO (red) cells were
treated with increasing concentrations of cytochalasin D for 2 h in complete culture
medium before fixation and staining. Images were analyzed and curves fitted using
nonlinear regression (mean ±SD, n = 4 (HeLa, NIH 3T3) or n = 8 (CHO)) for the
concentration range shown using GraphPad™ Prism software. IC50 values of 0.204,
1.084, and 1.011 µM were calculated for HeLa, CHO and NIH 3T3 cells respectively.
Dose response data for the treatment of NIH 3T3 cells with
increasing concentrations of staurosporine show a reduction in
fiber length demonstrated as a reduction in the maximum chord
straight values (Fig 5). An IC50 value of 0.095 µM was calculated
for staurosporine, which is consistent with literature reports of
inhibition of actin filament formation in the nanomolar range (20
to 50 nM) for this cell type (5). The exact mechanism of action of
staurosporine on the actin cytoskeleton is unclear. It is thought to
be partially attributed to its action as an inhibitor of protein kinase
C but could also involve other protein kinases (8).
Fig 6. Screen shot of Spotfire DecisionSite visualization of actin analysis. CHO
cells were treated with cytochalasin D dose response (n = 8 replicates).
Graphs shown include histogram of the distribution of lengths of actin
fibers—green, light blue, dark blue and red bars represent the column
position on the plate and therefore the concentration of cytochalasin D
corresponding to 0 to 1 nM, 3 to 27 nM, 82 to 740 nM, and 2.2 to 20 μM
respectively. The height of each bar represents the total number of fibers
within each specified length range. Scatter plot shows a dose response
curve based on fiber length per cell. Pie chart shows a plate map showing
fiber length per cell (by color) and maximum chord straight per cell (by size).
F-actin analysis screening assay
HeLa and CHO cells treated with 3 μM cytochalasin D for 2 h
prior to fixation and screening (n = 48 replicates per
treatment) produced good S:N and Z’ data (Table 1). HeLa and
CHO cells produced S:N values of 8.95 and 11.93 respectively
and values for Z’ were calculated to be 0.55 and 0.7 for the
same cell types respectively. These data demonstrate the
suitability of F-actin analysis for a screening assay.
Table 1. S:N and Z’ values for cell lines treated with cytochalsin D.
Fig 5. Dose response curve for the effect of staurosporine on F-actin fibers.
NIH 3T3 cells were treated with increasing concentrations of staurosporine
for 2 h in complete culture medium before fixation and staining. Images were
analyzed and curves fitted using nonlinear regression (mean ±SD, n = 4) using
GraphPad Prism software and an IC50 value of 0.095 µM was calculated.
HeLa
CHO
S:N
8.95
11.93
Z’
0.55
0.70
28-9327-13 AA
3
Conclusions
IN Cell Investigator software was utilized to quantitate disruption
of the actin cytoskeleton. The effects of cytochalasin D and
staurosporine on different cell types was investigated. The F-actin
structure within the cells was identified using Texas Red-X
phalloidin and the cytoskeleton was imaged using IN Cell
Analyzer 1000. Analysis of the images was achieved by
constructing a user-defined protocol using the Developer
Toolbox within IN Cell Investigator software.
The cytoskeleton was found to be organized as a mostly parallel
arrangement of linear actin fibers which became disrupted
following treatment with either cytochalasin D or staurosporine.
Cytochalasin D was shown to induce shortening of actin fibers in
all the cell types tested resulting in IC50 values of 0.204, 1.084, and
1.011 μM for HeLa, CHO, and NIH 3T3 cells respectively.
Treatment of NIH 3T3 fibroblasts with staurosporine caused the
defined linear actin staining to become more diffuse. An IC50 of
0.095 μM was calculated.
Visualization of data using Spotfire DecisionSite allowed a variety
of measures to be assessed for individual cells or populations.
Measures defining fiber or chord length provided robust
information relating to the actin cytoskeleton. Further
For contact information for your local office,
please visit, www.gelifesciences.com/contact
GE Healthcare Bio-Sciences Corp
800 Centennial Avenue
P.O. Box 1327
Piscataway
NJ 08855-1327
USA
www.gelifesciences.com/incell
examination of fiber length revealed that shorter fibers of F-actin
were predominant in wells corresponding to higher
concentrations of cytochalasin D (> 500 nM) whereas longer actin
fibers were only present at low concentrations of cytochalasin D
(< 500 nM) or in untreated cells. This agreed with the reported
action of cytochalasin D binding to the growing end of the actin
filament, inhibiting further elongation.
Analysis of the actin cytoskeleton and its reorganization due
to cytochalasin D treatment was shown to be suitable for a
screening assay. Assays set up for HeLa and CHO cells
demonstrated Z’ values of 0.55 and 0.7 respectively.
References
1. Carlier, M. F. et al. The Mechanisms of ATP Hydrolysis Accompanying the
Polymerization of Mg-actin and Ca-actin. J. Biol. Chem. 262, 3052–3059 (1987).
2. Dos Remedios, C. G. et al. Actin Binding Proteins: Regulation of Cytoskeletal
Microfilaments. Physiol. Rev. 83, 433–473 (2003).
3. Small, J. V. et al. The comings and goings of actin: coupling protrusion and
retraction in cell motility. Curr. Op. Cell. Biol. 17, 517–523 (2005).
4. Hall, A. Rho GTPases and the Actin Cytoskeleton. Science 279, 509–514 (1998).
5. Hedberg, K.K. et al. Staurosporine Induces Dissolution of Microfilament Bundles by
a Protein Kinase C-Independent Pathway. Exp. Cell Res. 188, 199–208 (1990).
6. Dubinsky, W. P. et al. Volume regulatory responses of basolateral membrane
vesicles from Necturus enterocytes: Role of the cytoskeleton. Proc. Natl. Acad. Sci.
USA 96, 9421–9426 (1999).
7. Cooper, J. A. Effects of Cytochalasin and Phalloidin on Actin. J. Cell Biol. 105,
1473–1478 (1987).
8. Yang, R. S. et al. Mechanism of the Morphological Changes Induced by
Staurosporine in Rat Osteoblasts. Calcif. Tissue Int. 61, 68–73 (1997).
GE, imagination at work, and GE monogram are trademarks of General Electric Company.
The IN Cell Analyzer 1000 is the subject of US patent application number 10/514925,
together with other pending family members, in the name of GE Healthcare Niagara,
Inc. The IN Cell Analyzer 1000 and associated analysis modules are sold under license
from Cellomics Inc. under US patent numbers US 5989835, 6416959, 6573039, 6620591,
6671624, 6716588, 6727071, 6759206, 6875578, 6902883, 6917884, 6970789, 6986993,
7060445, 7085765, 7117098 ; Canadian patent numbers CA 2282658, 2328194,
2362117, 2381334; Australian patent number AU 730100; European patent numbers EP
0983498, 1095277, 1155304, 1203214, 1348124, 1368689; Japanese patent numbers JP
3466568, 3576491 3683591 and other pending and foreign patent applications. All third
party trademarks are the property of their respective owners. © 2008 General Electric
Company—All rights reserved. First published Jan. 2008. All goods and services are sold
subject to the terms and conditions of sale of the company within GE Healthcare which
supplies them. A copy of these terms and conditions is available on request. Contact
your local GE Healthcare representative for the most current information. GE Healthcare
Bio-Sciences AB, Björkgatan 30, 751 84 Uppsala, Sweden. GE Healthcare UK Ltd,
Amersham Place, Little Chalfont, Buckinghamshire, HP7 9NA UK. GE Healthcare Europe
GmbH, Munzinger Strasse 5, D-79111 Freiburg, Germany. GE Healthcare Bio-Sciences KK,
Sanken Bldg. 3-25-1, Hyakunincho, Shinjuku-ku, Tokyo 169-0073 Japan.
28-9327-13 AA 01/2008