Chem*3560 Lecture 11: Regulation by proteolytic cleavage
advertisement

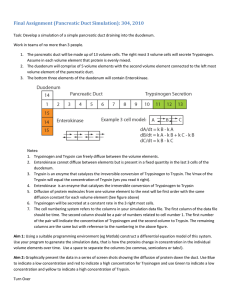
Chem*3560 Lecture 11: Regulation by proteolytic cleavage Some enzymes are synthesized in an inactive form called a zymogen or a proenzyme, which is only activated when the enzyme is needed for use (Lehninger p.286). Activation requires one or more key peptide bond(s) to be cut, which allows the enzyme's tertiary structure to rearrange so that the final active state can form. Activation by proteolytic cleavage is usually a one-time event - it’s not easy to rejoin the broken peptide chain to undo the change. Digestive enzymes A variety of digestive proteases are synthesized in the pancreas as inactive proenzymes, so that they don’t start to attack the cells that make them. Pancreatic cells are specialized as protein synthesis factories responsible for making and secretting the digestive proenzymes as well as the hormones glucagon and insulin. Digestive proenzymes are stored in the pancreatic cells as secretory granules, and empty into the duodenum via the pancreatic duct, whereas the hormones are secreted to the bloodstream. Proenzyme Enzyme Function Trypsinogen Chymotrypsinogen Procarboxypeptidase Proelastase Trypsin Chymotrypsin Carboxypeptidase Elastase cuts large polypeptides at Arg or Lys cuts large polypeptides at Phe, Tyr or Trp removes C-terminal amino acid from oligopeptides Specific for elastin The above enzymes are active at pH slightly above neutral, corresponding to conditions in the small intestine. In addition, pepsinogen is made in the stomach, and converted to pepsin, which is normally active at the very low pH in the stomach. Pepsin also acts on large polypeptides. Activation of trypsinogen Trypsinogen is activated by proteolytic cleavage (hydrolysis) of the peptide bond between Lys 6 and Ile 7. The oligopeptide with the first 6 amino acids is lost and Ile 7 becomes the new N-terminal. The positive charge associated with the N-terminal is now in a new position, where it can tuck into the protein core and ion-pair with Asp 194, which realigns the peptide loop containing Asp 194. (O atoms - red, N atoms - blue, C atoms - grey, H atoms omitted) The neighbours of Asp 194, Gly 193 and Ser 195, make up the oxyanion hole and the catalytic nucleophile in trypsin and chymotrypsin. The full length trypsinogen does not have the correct organization of the catalytic site, because the Ser 195 side chain is wrongly positioned relative to His 57 of the catalytic triad, and trypsinogen does not bind peptide in a way that guides substrate to form the hydrolysis transition state. The reaction that activates trypsinogen is induced by the protease enteropeptidase, which is a membrane protein exposed on the external surface of the mucosal cells that line the duodenum. Enteropeptidase is specific for the sequence Lys-Ile found in trypsinogen. The quantity of enteropeptidase present is very small, and may only activate a small fraction of the total trypsinogen. Trypsin is the common activator of the pancreatic proenzymes Since the target site in trypsinogen is a surface exposed -Lys-Ile-, trypsinogen is also a trypsin substrate. Trypsin that is initially made active by enteropeptidase can finish off the process of activating the remaining trypsinogen. This leads to an escalating rate of activation in the duodenum, a positive feedback effect. As trypsin gains activity, it is also responsible for activating chymotrypsinogen, procarboxypeptidase and elastase. Trypsin cuts chymotrypsinogen between Arg 15 and Ile 16, at a location which is structurally equivalent to Lys6 and Ile7 in trypsinogen. The newly exposed N-terminal Ile is similarly positioned to make an ion pair with Asp 194, and the mechanism of activation is the same as for trypsin. The immediate product is called π -chymotrypsin and is fully active as a protease. Self-exposure to π-chymotrypsin results in additional cleavages occurring at Leu13, Tyr146 and Asn148. The dipeptides Ser14-Arg15 and Thr147-Asn148 are lost, but the three large oligopeptides remain held together by disulfide bonds. The final product is α -chymotrypsin (Lehninger p.287). Trauma to the internal organs that damages the pancreas can cause pancreatitis, an unpleasant and painful condition in which traces of trypsin leak out and attack cells, activating their stored zymogen granules. The result is an escalating tendency of the pancreas to digest itself and surrounding organs. To prevent premature activation of trypsinogen, pancreas also produces small amounts of secretory pancreatic trypsin inhibitor. This is a small protein that exposes a substrate-like loop that is attacked by trypsin. However, the inhibitor is designed to bind trypsin extremely tightly (Ki = 10–13 M), so the reaction product is not released easily and continues to occupy the catalytic site. There is enough inhibitor to control any traces of trypsin that might develop accidentally, and prevent it from triggering the activation of the remaining trypsinogen while still in the pancreas. Once the trypsinogen is secreted into the duodenum, enteropeptidase can make enough trypsin to overcome the trace of inhibitor, and self activation can proceed. Pepsinogen is activated by change in the pH of its environment Pepsinogen is activated when it is secreted into the stomach. The intracellular environment has normal pH, but the stomach contents are at pH 2. The N-terminal segment of pepsinogen has 6 Arg and Lys, but the rest of the polypeptide is rich in Asp and Glu. As a result the N-terminal region binds tightly into the catalytic site through ion pairs, so that other substrates are excluded. In addition, the catalytic site of active pepsin consists of a pair of Asp side chains, one as -CO2 H to act as general acid and one as -CO2 – to act as general base. At neutral pH, both Asp side chains are deprotonated. When pepsinogen is secreted into the stomach, the low pH causes several carboxylate groups to become protonated, incuding the key Asp in the catalytic site, and this weakens the binding to the N-terminal segment. Pepsinogen starts by hydrolyzing itself to split off the N-terminal peptide of 44 amino acids, thereby opening up the catalytic site. Elastase is regulated by α 1-antitrypsin Elastase is secreted by white blood cells, and is responsible for cleaving elastin, a connective tissue protein. Elastin forms a molecular network that is highly cross-linked (Lehninger p.122), and by cutting the main polypeptide at intervals, the network becomes stretchable. This gives elastic properties to tissues such as lung, blood vessels and gut.. However, too much cleavage results in the elastin falling apart, so tissues lose elasticity. The correct degree of cleavage is maintained by balancing the action of elastase with α 1-antitrypsin, a serum protein that is misnamed. (Serum is the fluid component of blood that reamins after red and white cells are removed.) Although α 1-antitrypsin inhibits tryspin slightly, trypsin is not found in serum and the inhibitor really acts on elastase under physiological conditions. Elastase binds α1-antitrypsin in the region of Met 358. In smokers, Met can become oxidized to methionine sulfoxide, and the modified α1-antitrypsin does not inhibit elastase. The excessive action of elastase on lung tissue results in loss of lung elasticity and difficulties in breathing known as emphysema.