2-DE/MALDI-MS
advertisement

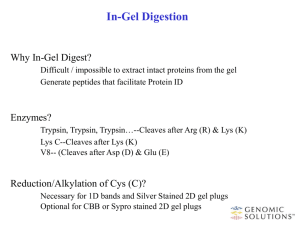
2-DE/MALDI-MS The identification of spots from 2-DE gels was described in detail [28]. After digestion the peptides were dissolved in 1.3 µl 33% ACN, 0.1% TFA; 0.25 µl of the solution was mixed with 0.5 µl 5 mg/ml alpha-cyano-4-hydroxycinnamic acid (Aldrich 1302B1, recrystallized and stored at -20°C), 50% ACN, 0.3% TFA and spotted onto the MALDI steel target. Protein spots that failed identification were further purified with ZipTipµ-C18 (Millipore, Schwalbach, Germany) according to the instructions of the manufacturer. The samples were analyzed in the MS mode for generation of peptide mass fingerprints (PMF) with a 4700 Proteomics Analyzer (Applied Biosystems, Darmstadt, Germany) [28]. Concomitantly, the peptides of the three most intense peaks were fragmented in the TOF/TOF mode to complement the protein identification by PMF. MS spectra were calibrated internally by trypsin fragments and known contaminants containing matrix, trypsin and keratin peptide masses (suppl. Material Table 1) were excluded by an exclusion list. Protein identification was performed using the GPS Explorer Software 3.5. and was achieved by database comparison against the H. pylori 26695 subset of the NCBI database using the search algorithm MASCOT 1.9. A peptide tolerance of 30 ppm, a MS/MS tolerance of 0.3 Da and one missed cleavage were allowed. Moreover, the variable modifications N-terminal acetylation, Met-oxidation, propionamide (C) and pyroglutamic-acid on N-terminal Gln were allowed. An exclusion list was generated (supplemental material Table2 excel) using MS-Screener [32], whereby mass values which occurred with a frequency of more than 5% and those which did not fulfil the half decimal place rule were excluded. Proteins were considered as identified, if (i) the sequence coverage was >30%, or (ii) the sequence coverage was > 10% and at least one MS/MS spectrum confirms identification and the MascotScore is >44 with p <0.05, or (iii) in some rare exceptions the first two criteria were not fulfilled, but the spot under investigation was within a series of identified spots or the mass loss rules verified an MS/MS assignment [33].