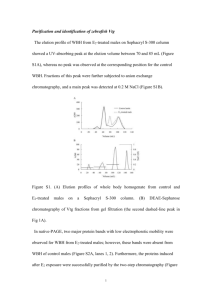
Inter-Laboratory Validation of a homologous ELISA to quantify
advertisement

Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA Inter-Laboratory Validation of a homologous ELISA to quantify Vitellogenin in the fathead minnow (Pimephales promelas) Janne K. Eidem, Hans Kleivdal and Anders Goksøyr Biosense Laboratories AS Thormøhlensgt 55 N-5008 Bergen http://www.biosense.com Contact: Janne K. Eidem +47 55543980 janne@biosense.com Page 1 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA TABLE OF CONTENTS SUMMARY INTRODUCTION The fathead minnow Vitellogenin ELISA Brief summary of method PURPOSE OF THE VALIDATION AIMS OF THE VALIDATION STUDY STUDY DESIGN Participant Laboratories Sample preparation and spiking Sample and test kit shipments Data reporting and handling VALIDATION RESULTS Applicability (Scope) Precision Between-Day Precision RSDr Between-Laboratory Precision RSDR Accuracy DISCUSSION CONCLUSION ACKNOWLEDGEMENTS REFERENCES AND LINKS APPENDIX A. Single-Lab Validation Report B. Letter to participants C. Study protocol and report sheet D. Result sheet E. Individual raw data Page 2 3 3 5 7 7 8 8 8 9 9 9 10 10 10 11 12 12 13 15 15 16 17 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA SUMMARY Vitellogenin (Vtg) is an established and sensitive endpoint for analysis of endocrine disruption in fish. The widespread use of Vtg in this regard has led to the need for standardised assays to quantify Vtg in fish samples. Fathead minnow (Pimephales promelas) is an important fish test species used in ecotoxicology laboratories across the world. Based on monoclonal antibodies raised against fathead minnow (FHM) Vtg, we have developed a quantitative sandwich enzyme-linked immunosorbent assay (ELISA) for FHM Vtg. A Single-Laboratory Validation has been performed earlier, according to international guidelines. Here, an Inter-Laboratory Validation study was carried out in order to determine the performance of the FHM Vtg ELISA in four different laboratories. Study samples consisted of both plasma, whole body homogenate (WBH) samples and ELISA kit Dilution buffer. The results are summarised in Table 1, and show that the FHM Vtg ELISA is suitable for quantification of Vtg in both plasma and WBH samples from FHM in different laboratories. Table 1: Summary of results from Inter-Laboratory Validation study of the FHM Vtg ELISA kit Performance characteristics Aim (%)1 Value (%) Between-Day Repeatability Precision, RSDr 16.4 Between-Lab Reproducibility Precision, RSDR >50 18.6 Recovery 50-200 69.4 Bias -30.6 1 Goksøyr et al 2003 *** INTRODUCTION Vitellogenin (Vtg) is a large phospholipoglycoprotein, which functions as the egg yolk precursor in oviparous vertebrates such as fish. The Vtg protein is produced in the liver and secreted from the liver cells through the secretory pathway, enters the blood circulation where it is transported to and taken up by growing oocytes. Endogenous oestrogen levels regulate Vtg production (Figure 1), and plasma Vtg concentrations normally indicate the maturational status of the female fish (for reviews, see Mommsen & Walsh, 1988; Arukwe & Goksøyr, 2003). More than a decade ago, several studies demonstrated that also male fish, caught in rivers and streams, had high levels of plasma Vtg (e.g. Purdom et al., 1994; Jobling et al., 1998), caused by chemicals present in the environment, acting like estrogens. Since then, numerous studies have shown the fish Vtg to be a highly responsive biomarker for estrogenic compounds in both in vitro hepatocyte cell cultures, in vivo aquaria studies, and field studies (for reviews, see Kime, 1995; Sumpter & Jobling, 1995; Arukwe & Goksøyr, 1998; 2003). Vtg induction in fish has now become an accepted measure of xenoestrogenic potency of chemicals, effluents, and discharges. Page 3 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA X Liver Vitellogenin Zona radiata P. Gonads Estradiol Figure 1: In response to oestradiol or xenoestrogens (x), Vitellogenin (egg yolk precursor) and zona radiata proteins (egg shell precursors) are produced in the liver and transported via the blood to the gonads. Within international bodies such as the Organization for Economic Cooperation and Development (OECD), work is ongoing to develop screening and testing programmes for endocrine disrupting effects of new chemicals. In the focus of this development are small fish test species including the fathead minnow (Pimephales promelas), zebrafish (Danio rerio), and Japanese medaka (Oryzias latipes). These fish share several attributes that make them ideal test species for reproductive toxicity testing, including small size at maturity, relatively short generation times, asynchronous spawning, and overall ease of culture. Against this background, there is a need for specific, sensitive, and reliable methods for measuring Vtg levels in these fish species, assays that are readily available and give reproducible results in different laboratories. The enzyme-linked immunosorbent assay (ELISA) is a sensitive laboratory technique widely used to detect and quantify antigens in a variety of biological samples. They can be quantitative (with a standard curve) or qualitative (semi-quantitative - without a standard curve). The two most widely used principles for quantitative detection of proteins are the competitive ELISA and the sandwich ELISA techniques (Crowther, 2001). *** Page 4 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA THE FATHEAD MINNOW VITELLOGENIN ELISA The Biosense fathead minnow (FHM) Vtg ELISA is based on a sandwich format, utilizing two different monoclonal mouse antibodies (Abs) raised against FHM Vtg. The antibodies were developed at the University of Florida, Gainesville, USA. Figure 2: The principle of the fathead minnow Vtg ELISA. The capture antibody is immobilised on ELISA microtiter plates, and binds to FHM Vtg in the standard or sample added to the ELISA well (Figure 2). After unbound components are washed away, a Detecting Ab, labelled with the enzyme horseradish peroxidase (HRP), is added. This Ab binds to a different part of the Vtg molecule, creating a sandwich of antibodies and Vtg. Addition of the HRP substrate Tetramethyl Benzidine (TMB) results in a colour reaction where the enzyme catalyses the conversion of this uncoloured substrate to a blue product. After development, the reaction is stopped by addition of a mild sulphuric acid, changing the colour from blue to yellow. The colour intensity is measured using a microplate reader with a 450 nm filter, and is proportional to the concentration of Vtg in the standard/sample. All absorbance levels are corrected for non-specific background reading (NSB), and a calibration curve is created by plotting Vtg concentration on the x-axis and the corresponding absorbance level on the y-axis (Figure 3). A 4 parameter or a log-log curve fit can be used to describe the relationship between the concentration and the signal. The equation for this calibration curve is then used to calculate the Vtg concentration in plasma or WBH samples. *** Page 5 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA FHM Vtg standard 100 10 90 80 70 60 50 0.1 40 %CV A450-NSB 1 30 0.01 20 10 0.001 0.01 0 0.1 1 10 100 ng/ml Standard Within-Day Precision Between-Day Precision Figure 3: Fathead minnow Vtg standard curve, compiled from 15 assays performed during the Single-Laboratory Validation (using Microsoft Excel). The secondary y-axis shows the Within-Day and Between-Day Repeatability Precisions (RSDr) for the standard curves. A Single-Laboratory Validation of the FHM Vtg ELISA was performed at Biosense Laboratories AS (Eidem et al 2005a). The validation demonstrated that the ELISA is both sensitive, precise and reliable and thus fit for its intended purpose. Figure 3 shows a representative standard curve derived from 15 assays performed during the Single-Lab Validation, including Between-Day and Within-Day Repeatibility Precision (RSDr) for the whole standard curve concentration range. Table 2 summarises the results of the Single-Laboratory Validation. The complete results from this validation are presented in Appendix A. Table 2: Summary of Single-Laboratory Validation results (Eidem et al 2005a) Performance characteristics Aim1 Value Selectivity Matrix blank < LOD No response at minimum (with the necessary dilution dilution = 1:50 (plasma), 1:100 factor to avoid matrix effects) (WBH) Calibration Standard curve working range Standard curve working range >10-fold, preferably 50-100 0.1-25 ng/ml (250-fold) fold to be practical with the dynamic range found in Vtg levels Accuracy (Recovery) Ideally 50-200% 75-106% Repeatability Precision RSDr <20% Within-Day RSDr: 4.5% Between-Day RSDr: 9.9% Limit of Detection (LOD) <10 ng/ml 0.02 ng/ml (plasma) 0.04 ng/ml (WBH) Limit of Quantification (LOQ) <10 ng/ml 0.09 ng/ml (plasma) 0.11 ng/ml (WBH) Sample LOQ 200 – 500 ng/ml 4.68 ng/ml (plasma, 1:50) (=LOQ x necessary matrix dilution) 11.35 ng/ml (WBH, 1:100) Comparison with Biosense R2>0.99 Carp Vtg ELISA kit Comparison with competitor R2>0.99 FHM Vtg ELISA kit Page 6 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA BRIEF SUMMARY OF METHOD Equipment and reagents required in addition to the FHM Vtg ELISA kit: • 0.3M H2SO4 (stop solution) • Microplate reader (wavelength 450 nm) • Pipettes with disposable tips (5-1000 µl) • Multi-channel pipette and reagent reservoir. Alternatively, a stepper pipette with disposable tips (100 µl) can be used. • Test tubes (1-50 ml) • Microplate washing device (an automatic or manual plate washer is recommended, but a squeeze bottle or a multichannel/stepper pipette can also be used) • Vortexer • Crushed ice Summary of the ELISA method: 1. Thaw samples on ice. 2. Prepare dilutions of standard and samples on ice. 3. To the pre-coated plates, add 100 µl Dilution buffer to the NSB wells. Add 100 µl of diluted standards and samples to the remaining wells. Incubate at room temperature for 1.5 hour. 4. Wash the plates three times with 200 µl Washing buffer per well. Add 100 µl of diluted Detecting antibody to all wells. Incubate at room temperature for 0.5 hour. 5. Wash the plates five times with 200 µl Washing buffer per well. Add 100 µl Substrate solution to all wells. Incubate in the dark at room temperature for 20 minutes. 7. Add 100 µl of 0.3M H2SO4 to all wells to stop the reaction. 8. Read the absorbance at 450 nm. 9. Calculate the results. Total sample capacity: 12 samples per plate (three dilutions of each sample, analysed in duplicates) Total assay time: 2 hours, 20 minutes. *** PURPOSE OF THE VALIDATION In the process of evaluating Vtg as an endpoint for endocrine disruptor testing and screening, various studies have been conducted over the recent years, involving both non-commercial laboratory methods and a few commercially available ELISA kits (for zebrafish Vtg inter-comparison study see http://abstracts.co.allenpress.com/pweb/setac2003/document/?ID=30012, for an overview of validation status in the US, see http://www.epa.gov/scipoly/oscpendo/assayvalidation/status.htm). These studies Page 7 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA have demonstrated the variance between different Vtg standards, between different assays and between different laboratories using the same assay. Clearly, there is a need for standardised, reproducible methods/kits that have been thoroughly characterised, with validation data describing the assay’s performance, thus giving the end user reasonable expectancies. The aim of the following study was to obtain inter-laboratory data on Precision and Accuracy, according to international guidelines (http://www.aoac.org). *** AIMS OF THE VALIDATION STUDY In a 2003 document, Biosense took the initiative to stress the need for standardised validations of Vtg standards and assays (Goksøyr et al, 2003). In this document, which was based on experience from participation in studies comparing Vtg methods, a set of performance criteria was suggested, to be met by Vtg quantification methods (see under “Aims” in Table 1). In this Inter-Laboratory Validation, a set of characteristics was analysed, chosen on the basis of international guidelines from the AOAC (http://www.aoac.org). The terminology involved in validation work is often confusing, and depends largely on which set of guidelines one looks at. Definitions used in this report is based on http://www.aoac.org/intaffairs/analytical_terminology.htm This study aimed to obtain Inter-Laboratory Validaion data on the following parameters: 1. Applicability (Scope) The analytes, matrices and concentrations for which a method of analysis may be used satisfactorily 2. Precision (Within-Laboratory, Between-Laboratory) Closeness of agreement between independent test results obtained under stipulated conditions. 3. Accuracy (Recovery, Bias) Closeness of agreement between a test result and an accepted reference value. *** STUDY DESIGN PARTICIPANT LABORATORIES In addition to Biosense Laboratories AS, three external laboratories were invited to participate in the validation. These laboratories have long, previous experience with the ELISA technique, removing the lack of experience factor from the results. *** Page 8 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA SAMPLE PREPARATION AND SPIKING Sample blanks: Plasma from male fathead minnows was purchased from Fish Soup, Newberry, Florida, USA. Plasma from 34 individual males was screened, and samples having non-detectable levels of Vtg at a 1:100 dilution were pooled. WBH from male fathead minnows was a gift from Robert Bringolf, Iowa State University, USA. The WBH showed no detectable levels of Vtg at a 1:100 dilution. Spiked samples: For spiking/recovery studies, Sample blanks (plasma, WBH and kit Dilution buffer) were diluted 10-fold with kit Dilution Buffer containing different concentrations of purified, non-lyophilised Vtg. Samples were mixed thoroughly, aliquoted into suitable volumes and frozen at –80oC. Concentrations in spiked samples were 5, 25 and 125 ng/ml. Naturally incurred samples: Plasma with low and medium levels of Vtg was purchased from Fish Soup, Newberry, Florida, USA. Plasma from oestradiol (E2)-treated fish, as well as WBH with low and medium levels of Vtg was a gift from Robert Bringolf, Iowa State University, USA. WBH from E2-treated FHM was prepared at Biosense Laboratories. Samples were mixed thoroughly, aliquoted into suitable volumes and frozen at –80oC. *** SAMPLE AND TEST KIT SHIPMENT Aliquoted samples were shipped on dry ice using a courier company. Upon arrival, the participating laboratories verified that there was dry ice left in the container and that the samples were in good (frozen) condition and that they were transferred to –80oC until analysis (see Report Sheet in Appendix C). The ELISA kits were shipped on cool packs (not frozen) and were transferred to 4oC upon arrival. *** DATA REPORTING AND HANDLING Each participating laboratory was given detailed instructions, plate layout and a report sheet into which the plate reader raw data was placed (see Result Sheet, Appendix C). The data handling was checked with respect to the following (by study leader at Biosense Laboratories AS): 1. Standard curves were adjusted according to kit protocol. Page 9 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA 2. Each standard and sample dilution had been performed in triplicates. If the CV between triplicate absorbance values was > 10%, the following was done: • • identify and remove a clear outlier to reduce CV to < 10% if no clear outlier was identifiable, the whole data point was excluded 3. Sample dilutions with absorbance levels over or under the defined standard curve working range were discarded. 4. If more than one sample dilution fell within the working range of the standard curve, the average of these was calculated. If the results from these different dilutions had a CV>50%, this data point was excluded. *** VALIDATION RESULTS APPLICABILITY (SCOPE) Applicability (Scope): The analytes, matrices and concentrations for which a method may be used satisfactorily. This Inter-Laboratory Validation was performed in order to validate the performance of the FHM Vtg ELISA, developed for use in endocrine disruptor screening tests. The same material was analysed by four different laboratories with previous ELISA skills. The ability of the ELISA to accurately analyse Vtg levels in spiked (fortified) plasma, WBH and the ELISA kit Dilution buffer was analysed. Four levels of Vtg were used in this validation (0, 0.5, 2.5 and 12.5 ng/ml Vtg), corresponding to different levels within the standard curve working range (0.1-25 ng/ml). The following validation data demonstrate that the FHM Vtg ELISA kit is suitable for Vtg analysis in the matrices and in the concentrations ranges included in the validation. *** PRECISION Precision is the closeness of agreement between test results obtained under stipulated conditions. • Repeatability Precision (same laboratory and operator, samples, equipment, short time intervals), separated into Within-Day and Between-Day Repeatability precision, usually expressed as relative standard deviation, RSDr. • Reproducibility Precision (different laboratories, equipment and operators, same samples). Usually expressed as relative standard deviation, RSDR. Spiked and unspiked plasma and WBH sample blanks, as well as the ELISA kit Dilution buffer, were analysed. On two different days, one aliquot was thawed, diluted 1:10 and analysed in triplicates. Unspiked plasma and WBH samples were analysed at three different dilutions according to kit protocol (plasma 1:50, 1:5000 and 1:500000; WBH 1:100, 1:10000 and 1:1000000). Page 10 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA Table 3 shows the compiled data from all four laboratories and both days as well as Between-Day Repeatability Precision RSDr. *** Between-Day Repeatability Precision RSDr: RSDr: Variation between results obtained for a given sample on the two different days. Average RSDr: Average RSDr for all samples analysed by one laboratory Overall RSDr: Average RSDr for all samples and laboratories Table 3: Between-Day Repeatability Precision RSDr Results from four laboratories, showing measured Vtg levels (ng/ml) and RSDr (%). Lab1 code Day 1 Lab2 Day 2 Average RSD Day 1 PJ <w.r. <w.r. - PQ 0.4 0.4 PB 2.0 PS WJ r Lab3 Day 2 Average RSD Day 1 - <w.r. <w.r. - 0.4 2.1 0.3 0.3 2.0 2.0 1.9 1.4 10.0 10.5 10.3 3.1 <w.r. <w.r. - - WQ 0.3 0.3 0.3 WB 1.5 1.4 WS 7.7 7.6 BJ <w.r. BQ 0.4 BB r Lab4 Day 2 Average RSD Day 1 - <w.r. <w.r. - 0.3 1.8 0.4 0.4 1.6 1.5 10.0 2.1 8.5 10.4 9.4 <w.r. <w.r. - 13.3 0.2 0.3 1.5 5.4 1.2 7.7 1.3 7.4 <w.r. - - 0.4 0.4 1.9 2.0 1.9 2.0 BS 10.5 10.9 PM 952 1180 PA 38642 54454 PR 3940983 4883251 4412117 WM 7.92 <w.r. WA 27.0 WR 469214 r Day 2 Average RSD r - <w.r. <w.r. - 0.4 6.4 0.5 nd 0.5 - 2.1 2.1 0.8 nd 1.6 1.6 - 14.5 13.0 12.5 12.7 2.8 8.1 10.1 9.1 15.7 - <w.r. <w.r. - - 1.0 <w.r. 1.0 - 0.2 17.7 <w.r. <w.r. - - <w.r. 0.2 0.2 - 1.2 1.2 0.1 1.0 1.2 1.1 13.6 1.7 nd 1.7 - 8.0 7.7 5.4 6.1 6.7 6.4 6.8 7.1 5.5 6.3 17.9 <w.r. <w.r. - - <w.r. <w.r. - - <w.r. <w.r. - - 0.3 0.3 0.3 17.5 0.4 0.4 0.4 11.8 nd <w.r. - - 5.2 1.8 1.8 1.8 1.7 2.2 1.9 2.1 8.2 <w.r. nd - - 10.7 2.6 10.4 11.2 10.8 4.9 12.1 11.9 12.0 1.5 5.2 11.0 8.1 50.9 1066 15.2 1124 919 1021 14.2 1574 1192 1383 19.6 2289 849 1569 64.9 46548 24.0 40192 37687 38939 4.6 57282 56422 56852 1.1 nd nd - - 15.1 4084024 4374596 4229310 4.9 5273682 5116503 5195092 2.1 3935890 >w.r. 3935890 - 7.92 - <w.r. <w.r. - - <w.r. <w.r. - - nd <w.r. - - 22.4 24.7 13.3 26.3 13.1 19.7 47.1 <w.r. <w.r. - - nd <w.r. - - 516990 493102 6.9 728601 603104 29.4 618288 478045 548166 18.1 nd Average RSDr 7.9 477607 12.4 409530 409530 7.7 Overall RSDr for all laboratories: - - 37.4 16.4 nd: not determined due to error or too high variance >w.r. and < w.r.: absorbance levels above or below working range of standard curve, respectively Table 3 shows that the overall variation RSDr between results obtained on two different days varied between 7.7 and 37.4% for the different laboratories. The overall RSDr for all four laboratories was 16.4%. Note that for Lab 4 RSDr was only obtainable for four samples. *** Page 11 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA Between-Laboratory Reproducibility Precision RSDR: RSDR: Variation between results obtained by different laboratories for one sample (average of two days). Overall RSDR : Average RSDR for all laboratories and samples Table 4: Between-Laboratory Reproducibility Precision RSDR. Results from four laboratories, showing measured Vtg levels (ng/ml, average of two days) and RSDR (%) code Average day1-2, Average day1-2, Average day1-2, Average day1-2, Average Lab1 Lab2 Lab3 Lab4 Lab1-4 PJ PQ 0.4 0.3 0.4 0.5 0.4 PB 2.0 1.5 2.1 1.6 1.8 PS 10.3 9.4 12.7 9.1 10.4 WJ 1.0 WQ 0.3 0.2 0.3 WB 1.5 1.2 1.1 1.7 1.3 WS 7.7 7.7 6.4 6.3 7.0 BJ BQ 0.4 0.3 0.4 0.4 BB 2.0 1.8 2.1 1.9 BS 10.7 10.8 12.0 8.1 10.4 PM 1066 1021 1383 1569 1260 PA 46548 38939 56852 47446 PR 4412117 4229310 5195092 3935890 4515561 WM 7.9 7.9 WA 24.7 19.7 22.2 WR 493102 603104 548166 409530 528325 Overall Between-Laboratory RSDR RSD Lab 1-4 18.5 16.7 16.3 15.0 17.7 12.3 15.6 7.4 21.0 37.6 20.0 12.6 28.7 20.6 18.6 R “-“ indicates that data was not obtainable for this sample (see Table 3) Table 4 shows that the variation RSDR between results obtained in four different laboratories varied between 7.4 and 37.6% for the different samples. The overall RSDR for all samples was 18.6%. *** ACCURACY Accuracy (Trueness) is the closeness of agreement between a test result and an accepted reference value. Recovery is the proportion of the amount of analyte, present in or added to, the analytical portion, which is extracted and presented for measurement. Bias is the difference between the test results and an accepted reference value. Recovery and Bias in spiked samples were determined using the following formulas: Recovery = (C1-C2)/C3 x 100 Where C1= concentration measured in spiked sample, C2= concentration measured in unspiked sample, C3= theoretical concentration. Page 12 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA Bias = (C3-(C1-C2))/C3 x 100 Where C1= concentration measured in spiked sample, C2= concentration measured in unspiked sample, C3= theoretical concentration Table 5: Recovery and Bias. Results from analysis of spiked samples, showing measured Vtg levels (ng/ml, average of four laboratories), Recovery (%) and Bias (%) Sample type and code Theoretic conc. (ng/ml) Average Lab1-4 (ng/ml) Recovery Lab 1-4 (%) Plasma 0 0.5 2.5 12.5 0 0.5 2.5 12.5 0 0.5 2.5 12.5 0.4 1.8 10.4 1.0 0.3 1.3 7.0 0.4 1.9 10.4 74.8 73.3 83.0 51.7 52.3 56.1 72.7 77.7 83.2 WBH Buffer PJ PQ PB PS WJ WQ WB WS BJ BQ BB BS Overall Recovery Bias 69.4 -30.6 “-“ indicates that data was not obtainable for this sample (see Table 3) The results show that the recovery and bias varied somewhat with both sample type and spike concentration, with an overall Recovery of 69.4%. See Discussion for more on this. One lab reported a false positive on one day (sample WJ, Lab 4, see Table 3). *** DISCUSSION An Inter-Laboratory Validation was performed to validate the performance of the Biosense FHM Vtg ELISA in four different laboratories. A set of 18 plasma, WBH and Buffer samples, comprising both spiked and unspiked samples, was analysed twice, on two different days. Variation within laboratories (between days) and between laboratories was analysed, as well as the ability of the assay to measure correct values in spiked samples. Kits and samples were successfully received and handled by participants, and data sets were obtained from all four laboratories. Samples analysed on two different days determined the Between-Day Repeatability Precision (RSDr). The RSDr varied from 7.7-37.4% for the different laboratories, with an overall RSDr of 16.4%. This value was elevated by a high variation and a low Page 13 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA number of RSDr values (only from four samples) from Lab 4. If the RSDr values of this laboratory are excluded, the overall RSDr is reduced to 9.4%. Samples analysed by different laboratories allowed the determination of BetweenLaboratory Reproducibility Precision (RSDR). The RSDR varied between 7.4 and 37.6% for the different samples, and the overall RSDR for all samples was 18.6%. Both the RSDr and the RSDR values are within the aims by Goksøyr et al (2003): RSDr < 20% and RSDR < 50%. Analysis of spiked samples allowed the calculation of Recovery and Bias. plasma, WBH and ELISA kit Dilution buffer was spiked to three different concentrations corresponding to low, medium and high concentration levels of the standard curve (working range = 0.1-25 ng/ml, see Single-Laboratory Validation, Appendix A/Eidem et al, 2005a). The Recovery varied somewhat with sample type and spike level: with Dilution buffer giving the highest and WBH samples giving the lowest Recovery. Higher spike concentrations gave somewhat higher Recovery. An overall Recovery of 69.4% was obtained, averaged over all sample concentrations and types. This value is within the suggested aim of 50-200% Recovery (Goksøyr et al. 2003). Vtg is an unstable molecule, prone to degradation (Arukwe and Goksøyr 2003). For this reason, spiking of samples should ideally be done on the day of analysis to avoid freeze-thaw cycles and degradation. However, in order to obtain data for determination of Between-Day and Between-Laboratory Precision, samples had to be diluted, aliquoted and frozen. This is likely to have affected the Recovery of Vtg in spiked samples. Spiking/recovery experiments performed in the Single-Laboratory Validation, without freezing and thawing of the spiked samples, indicate recovery in both plasma and WBH between 79-106% (see Table 2 and Single-Laboratory Validation, Appendix A/Eidem et al, 2005a). Some of the laboratories (especially Lab 4, but also Lab 2) reported high variance between replicate wells, caused by contamination of wells with enzyme-labelled Detecting Ab and a resulting unspecific colour development. Generally, one or more of several factors can cause high variation between replicate wells: • • • Imprecise pipettes and/or pipetting techniques Imprecise washing techniques, or, in case of automatic plate washers, uneven dispensing/aspiration Careless handling of plates and plate sealers, which may result in splashing and/or well-to-well contamination Variation between replicate analyses of the same sample may often have the same reasons as variation between replicate wells. Variation is reduced by ensuring consistent techniques when preparing standards, sample and antibody dilutions, when adding the solutions and when performing washing step. Also, keeping temperatures and incubation times consistent is important. These factors should be Page 14 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA relatively easy to overcome with practise and proper maintenance of pipettes and equipment. In addition to variation arising during performance of the assay, some variation is also inherent in the assay itself. There will be some variation between different plates, between different vials of Vtg standard RM and between different batches. However, these factors are low due to extensive quality controls during production. Low absorbance was reported in obe case (Day 1, Lab 4), when a different washing buffer than the one enclosed in the kit had been used – this wash buffer contained Na-azide as preservant, and which is known to inhibit HRP. On Day 2 the kit wash buffer was used, and higher absorbance was achieved. *** CONCLUSION A successful Inter-Laboratory Validation was performed on the FHM Vtg ELISA kit. The results show that the kit performs with low variation and with good accuracy and that it is fit for its intended purpose, namely reliable quantification of Vtg in plasma or WBH samples from the fathead minnow (Pimephales promelas). *** ACKNOWLEDGEMENTS Biosense Laboratories AS would like to thank the following participants for donating their time and for their valuable input on the assay (in alphabetical order based on institution): Dr. Nancy Denslow and Dr. Kevin J. Kroll University of Florida, Gainesville, US Phone: (352)-392-4700 Fax: (352)-392-4707 E-mail: kkroll@ufl.edu Dr. Grace Panter AstraZeneca, Brixham Environmental Laboratory Brixham, UK Phone: 01803 884261 Fax: 01803882974 E-mail: grace.panter@brixham.astrazeneca.com Dr. Charles Tyler and Dr. Ronny van Aerle University of Exeter Exeter, UK Phone: 01392 264450 Fax: 01392 263700 E-mail: C.R.Tyler@exeter.ac.uk, R.Van-Aerle@exeter.ac.uk Page 15 *** Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA REFERENCES AND LINKS AOAC “Harmonisation of analytical terminology in accordance with international standards” http://www.aoac.org/intaffairs/analytical_terminology.htm) Arukwe A, Goksøyr A (1998). Xenobiotics, xenoestrogens and reproduction disturbances in fish. Sarsia 83:225-241. Arukwe A, Goksøyr A (2003). Eggshell and egg yolk proteins in fish: hepatic proteins for the next generation. Oogenetic, population, and evolutionary implications of endocrine disruption. Comp Hepatol 2: 4. Brion F, Nilsen BM, Eidem JK, Goksøyr A, Porcher JM (2002). Development and validation of an enzyme-linked immunosorbent assay to measure vitellogenin in the zebrafish (Danio rerio). Environ Toxicol Chem 28: 1699-1708. Crowther JR (2001). The ELISA Guidebook. In: Methods Mol Biol 149. Humana Press, Totowa, NJ Eidem JK, Kleivdal HK, Goksøyr A (2005a) Single-Laboratory Validation of a homologous ELISA to quantify Vitellogenin in the fathead minnow (Pimephales promelas) (http://www.biosense.com/render.asp?ID=71&segment=3&session) Goksøyr A. et al (2003) On the need for a standardized set-up for validation studies of fish vitellogenin assays as an endpoint in endocrine disruptor testing and screening – a proposal. http://www.biosense.com/Docs/GoksoyrEtal2003.pdf Jobling S, Nolan M, Tyler CR, Brighty G, Sumpter JP (1998). Widespread sexual disruption in wild fish. Environ Sci Technol 32: 2498–2506. Kime DE (1995). The effects of pollution on reproduction in fish. Rev Fish Biol Fisheries 5:52-96 Mommsen TP, Walsh PJ (1988). Vitellogenesis and oocyte assembly. In: Hoar WS, Randall VJ (eds) Fish Physiology XIA. Academic Press, New York, pp 347-406. Nilsen B.M. et al 2004. Development of quantitative vitellogenin-ELISAs for fish test species used in endocrine disruptor screening. Anal Bioanal Chem 378:621 http://www.springerlink.com, DOI 10.1007/s00216-003-2241-2 Norberg B, Haux C (1985). Induction, isolation and a characterization of the lipid content of plasma vitellogenin from two Salmo species: rainbow trout (Salmo gairdneri) and sea trout (Salmo trutta). Comp Biochem Physiol 81B:869–876. Porcher J-M. et al (2003). Intercomparison of zebrafish vitellogenin quantification methods. Society of Environmental Toxicology and Chemsitry, 24th annual meeting North America, http://abstracts.co.allenpress.com/pweb/setac2003/document/?ID=30012 Purdom CE, Hardiman PA, Bye VJ, Eno NC, Tyler CR, Sumpter JP (1994). Estrogenic effects of effluents from sewage treatment works. Chem Ecol 8: 275-285. Silversand C, Haux C (1995). Fatty acid composition of vitellogenin from four teleost species. J Comp Physiol 164B: 593-599. Sumpter JP, Jobling S (1995). Vitellogenesis as a biomarker for estrogenic contamination of the aquatic environment. Environ Health Perspect 103 (Suppl 7): 173-178. US EPA Endocrine Disruptor Screening Program, Assay status table. http://www.epa.gov/scipoly/oscpendo/assayvalidation/status.htm *** Page 16 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA APPENDIX A: SINGLE-LAB VALIDATION REPORT Single-Laboratory Validation of a homologous ELISA to quantify Vitellogenin in the fathead minnow (Pimephales promelas) Janne K. Eidem, Hans Kleivdal and Anders Goksøyr Biosense Laboratories AS Thormøhlensgt 55 N-5008 Bergen http://www.biosense.com Contact: Janne K. Eidem +47 55543980 janne@biosense.com Page 17 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA TABLE OF CONTENTS SUMMARY INTRODUCTION The fathead minnow Vitellogenin ELISA Brief summary of method PURPOSE OF THE VALIDATION AIMS OF THE VALIDATION STUDY Definitions VITELLOGENIN STANDARD Source for RM purification Purification of RM Quality assurance of RM Quantification of RM Stabilization of RM SAMPLE PREPARATION AND SPIKING VALIDATION RESULTS Applicability (Scope) Calibration LoD and LoQ (sensitivity) Selectivity (matrix effect) Repeatability Precision (intra- and inter-assay variation) Accuracy Ruggedness Comparison with existing methods DISCUSSION CONCLUSION REFERENCES AND LINKS Page 18 3 4 5 6 7 7 8 9 10 10 11 12 13 14 15 15 15 16 18 19 22 23 25 27 28 29 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA SUMMARY Vitellogenin (Vtg) is an established and sensitive endpoint for analysis of endocrine disruption in fish. The widespread use of Vtg in this regard has led to the need for standardised assays to quantify Vtg in fish samples. Fathead minnow (Pimephales promelas) is an important fish test species used in ecotoxicology laboratories across the world. Based on monoclonal antibodies raised against fathead minnow (FHM) Vtg, we have developed a quantitative sandwich enzyme-linked immunosorbent assay (ELISA) for FHM Vtg. A Single-Laboratory Validation was performed according to international guidelines. Study samples consisted of both plasma, whole body homogenate (WBH) and ELISA kit Dilution buffer. The results are summarised in Table 1, and show that the FHM Vtg ELISA is suitable for quantification of Vtg in both plasma and WBH samples from FHM. Table 1: Summary of results from Single-Laboratory Validation results Performance characteristics Aim1 Selectivity Matrix blank < LOD (with the necessary dilution factor to avoid matrix effects) Calibration Standard curve working range >10-fold, preferably 50-100 fold to be practical with the dynamic range found in Vtg levels Accuracy (Recovery) Ideally 50-200% Repeatability Precision RSDr <20% Limit of Detection (LOD) <10 ng/ml Limit of Quantification (LOQ) <10 ng/ml Sample LOQ 200 – 500 ng/ml (=LOQ x necessary matrix dilution) Comparison with Biosense Carp Vtg ELISA kit Comparison with competitor FHM Vtg ELISA kit 1) Goksøyr et al 2003 Value No response at minimum dilution = 1:50 (plasma), 1:100 (WBH) Standard curve working range 0.1-25 ng/ml (250-fold) 75-106% Within-Day RSDr: 4.5% Between-Day RSDr: 9.9% 0.02 ng/ml (plasma) 0.04 ng/ml (WBH) 0.09 ng/ml (plasma) 0.11 ng/ml (WBH) 4.68 ng/ml (plasma, 1:50) 11.35 ng/ml (WBH, 1:100) R2>0.99 R2>0.99 *** INTRODUCTION Vitellogenin (Vtg) is a large phospholipoglycoprotein, which functions as the egg yolk precursor in oviparous vertebrates such as fish. The Vtg protein is produced in the liver and secreted from the liver cells through the secretory pathway, enters the blood circulation where it is transported to and taken up by growing oocytes. Endogenous oestrogen levels regulate Vtg production (Figure 1), and plasma Vtg Page 19 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA concentrations normally indicate the maturational status of the female fish (for reviews, see Mommsen & Walsh, 1988; Arukwe & Goksøyr, 2003). More than a decade ago, several studies demonstrated that also male fish, caught in rivers and streams, had high levels of plasma Vtg (e.g. Purdom et al., 1994; Jobling et al., 1998), caused by chemicals present in the environment, acting like estrogens. Since then, numerous studies have shown the fish Vtg to be a highly responsive biomarker for estrogenic compounds in both in vitro hepatocyte cell cultures, in vivo aquaria studies, and field studies (for reviews, see Kime, 1995; Sumpter & Jobling, 1995; Arukwe & Goksøyr, 1998; 2003). Vtg induction in fish has now become an accepted measure of xenoestrogenic potency of chemicals, effluents, and discharges. X Liver Vitellogenin Zona radiata P. Gonads Estradiol Figure 1: In response to oestradiol or xenoestrogens (x), Vitellogenin (egg yolk precursor) and zona radiata proteins (egg shell precursors) are produced in the liver and transported via the blood to the gonads. Within international bodies such as the Organization for Economic Cooperation and Development (OECD), work is ongoing to develop screening and testing programmes for endocrine disrupting effects of new chemicals. In the focus of this development are small fish test species including the fathead minnow (Pimephales promelas), zebrafish (Danio rerio), and Japanese medaka (Oryzias latipes). These fish share several attributes that make them ideal test species for reproductive toxicity testing, including small size at maturity, relatively short generation times, asynchronous spawning, and overall ease of culture. Against this background, there is a need for specific, sensitive, and reliable methods for measuring Vtg levels in these fish species, assays that are readily available and give reproducible results in different laboratories. The enzyme-linked immunosorbent assay (ELISA) is a sensitive laboratory technique widely used to detect and quantify antigens in a variety of biological samples. They can be quantitative (with a standard curve) or qualitative (semi-quantitative - without a standard curve). The two most widely used principles for quantitative detection of proteins are the competitive ELISA and the sandwich ELISA techniques (Crowther, 2001). We have previously developed quantitative sandwich Vtg ELISAs for zebrafish and Japanese medaka, as well as a homologous carp Vtg ELISA, which also works well Page 20 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA for fathead minnow Vtg (Nilsen et al 2003). Here, we present Single-Laboratory (inhouse) Validation data on a new, homologous quantitative sandwich ELISA for measuring Vtg in plasma and whole body homogenate samples of the fathead minnow. The validation has been carried out at Biosense, according to international guidelines for Single-Laboratory Validation (AOAC International Training Course, 2003; Thompson et al, 2002; Eurachem, 1998). *** THE FATHEAD MINNOW VITELLOGENIN ELISA The fathead minnow (FHM) Vtg ELISA is based on a sandwich format, utilizing two different monoclonal mouse antibodies (Abs) raised against FHM Vtg. The antibodies were developed at the University of Florida, Gainesville, USA. Figure 2: The principle of the fathead minnow Vtg ELISA. The capture antibody is immobilised on ELISA microtiter plates, and binds to FHM Vtg in the standard or sample added to the ELISA well (Figure 2). After unbound components are washed away, a Detecting Ab, labelled with the enzyme horseradish peroxidase (HRP), is added. This Ab binds to a different part of the Vtg molecule, creating a sandwich of antibodies and Vtg. Addition of the HRP substrate Tetramethyl Benzidine (TMB) results in a colour reaction where the enzyme catalyses the conversion of this uncoloured substrate to a blue product. After development, the reaction is stopped by addition of a mild sulphuric acid, changing the colour from blue to yellow. The colour intensity is measured using a microplate reader with a 450 nm filter, and is proportional to the concentration of Vtg in the standard/sample. All absorbance levels are corrected for non-specific background reading (NSB), and a calibration curve is created by plotting Vtg concentration on the x-axis and the corresponding absorbance level on the y-axis (Figure 3). A 4 parameter or a log-log curve fit can be used to describe the relationship between the concentration and the signal. The equation for this calibration curve is then used to calculate the Vtg concentration in plasma or homogenate samples. Page 21 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA y = 0.1381x1.012 R2 = 0.9988 A450-NSB 10 1 0.1 0.01 0.1 1 10 100 ng/ml Vtg Figure 3: Fathead minnow Vtg standard curve *** BRIEF SUMMARY OF METHOD Equipment and reagents required in addition to the FHM Vtg ELISA kit: • 0.3M H2SO4 (stop solution) • Microplate reader (wavelength 450 nm) • Pipettes with disposable tips (5-1000 µl) • Multi-channel pipette and reagent reservoir. Alternatively, a stepper pipette with disposable tips (100 µl) can be used. • Test tubes (1-50 ml) • Microplate washing device (an automatic or manual plate washer is recommended, but a squeeze bottle or a multichannel/stepper pipette can also be used) • Vortexer • Crushed ice Summary of the ELISA method: 1. Thaw samples on ice. 2. Prepare dilutions of standard and samples on ice. 3. To the pre-coated plates, add 100 µl Dilution buffer to the NSB wells. Add 100 µl of diluted standards and samples to the remaining wells. Incubate at room temperature for 1.5 hour. 4. Wash the plates three times with 300 µl Washing buffer per well. Add 100 µl of diluted Detecting antibody to all wells. Incubate at room temperature for 0.5 hour. 5. Wash the plates five times with 300 µl Washing buffer per well. Add 100 µl Substrate solution to all wells. Incubate in the dark at room temperature for 20 minutes. 7. Add 100 µl of 0.3M H2SO4 to all wells to stop the reaction. 8. Read the absorbance at 450 nm. 9. Calculate the results. Page 22 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA Total sample capacity: 12 samples per plate (three dilutions of each sample, analysed in duplicates) Total assay time: 2 hours, 20 minutes. *** PURPOSE OF THE VALIDATION In the process of evaluating Vtg as an endpoint for endocrine disruptor testing and screening, various studies have been conducted over the recent years, involving both non-commercial laboratory methods and a few commercially available ELISA kits (for zebrafish Vtg inter-comparison study see http://abstracts.co.allenpress.com/pweb/setac2003/document/?ID=30012, for an overview of validation status in the US, see http://www.epa.gov/scipoly/oscpendo/assayvalidation/status.htm). These studies have demonstrated the variance between different Vtg standards, between different assays and between different laboratories using the same assay. Clearly, there is a need for standardised, reproducible methods/kits that have been thoroughly characterised, with validation data describing the assay’s performance, thus giving the end user reasonable expectancies. The aim of the following study was to obtain Single-Laboratory Validation data, according to international guidelines (AOAC, Eurachem, IUPAC). *** AIMS OF THE VALIDATION STUDY In a 2003 document, Biosense took the initiative to stress the need for standardised validations of Vtg standards and assays (Goksøyr et al, 2003). In this document, based on experience from participation in studies comparing Vtg methods, we suggested a set of performance criteria to be met by Vtg quantification methods (Table 2). In this Single-Laboratory Validation (SLV), a set of characteristics were analysed, chosen on the basis of guidelines set up by international bodies like AOAC, Eurachem and IUPAC. It is recommended that “Single-Laboratory validation requires the laboratory to select appropriate characteristics for evaluation from the following: applicability, selectivity, calibration, accuracy, precision, range, limit of quantification, limit of detection, sensitivity and ruggedness” (Thompson et al 2002). Page 23 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA Table 2 Pre-determined performance criteria (Goksøyr et al 2003) Performance characteristics Aim Selectivity Matrix blank < LOD (with the necessary dilution factor to avoid matrix effects) Calibration Standard curve working range >10-fold, preferably 50-100 fold to be practical with the dynamic range found in Vtg levels Accuracy1) Ideally 50-200% Repeatability 2) <20% Reproducibility3) <50% Limit of Detection (LOD) <10 ng/ml Limit of Quantification (LOQ) <10 ng/ml Sample LOQ 200 – 500 ng/ml (=LOQ x necessary matrix dilution) 1) Referred to in this document as Recovery Referred to in this document Within-Day and Between-Day Repeatability Precision RSDr 3) Referred to in this document Between-Laboratory Reproducibility Precision RSDR 2) This study aimed to obtain Single-Laboratory Validation data on the following parameters: 4. Applicability 5. Calibration 6. Limit of detection (LOD), Limit of Quantification (LOQ) 7. Selectivity 8. Precision (Within-Day, Between-Day) 9. Accuracy (spiking/recovery, bias) 10. Ruggedness In addition, comparisons with existing methods were also conducted. *** DEFINITIONS The terminology involved in validation work is often confusing, and depends largely on which set of guidelines one looks at. The following definitions have been used in this report (see http://www.aoac.org/intaffairs/analytical_terminology.htm): Applicability (Scope): The analytes, matrices and concentrations for which a method may be used satisfactorily. Selectivity: The ability to measure accurately the analyte (Vtg) in the presence of components that may be expected to be present in the matrix (plasma and whole body homogenate). Calibration, is the empirical determination of the relationship between the parameter measured (e.g. ELISA absorbance) and the analyte (Vtg) concentration. The range of concentrations of analyte where such relationship is established is often referred to as "calibration range" or the "standard curve working range". Accuracy is the closeness of agreement between a test result and the accepted reference value of the property being measured. Page 24 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA Recovery is the proportion of the amount of analyte, present in or added to, the analytical portion, which is extracted and presented for measurement Bias is the difference between the test results and an accepted reference value Precision is the closeness of agreement between test results obtained under stipulated conditions. • Repeatability Precision (same laboratory and operator, samples, equipment, short time intervals), separated into Within-Day and Between-Day Repeatability precision, usually expressed as relative standard deviation, RSDr. • Reproducibility Precision (different laboratories, equipment and operators, same samples). Usually expressed as relative standard deviation, RSDR. Limit of Detection is the smallest amount or concentration that can be reliably distinguished from zero. Defined here as reagent blank + 3x standard deviation of reagent blank. Indicates that the analyte is present, but not necessarily allowing exact quantification. Limit of Quantification A concentration above which the analytical method can operate with an acceptable precision. Defined here as reagent blank + 10x standard deviation of reagent blank. Sample LoQ. The LoQ corrected for minimum dilution factor necessary to avoid matrix effect. Ruggedness. The ability of the measurement process to resist changes in results when subjected to minor changes in environmental and procedural variables. *** VITELLOGENIN STANDARD A validation of a quantitative Vtg ELISA should address not only the assay itself, but also the Vtg standard used. Here we describe the purification, quality assurance and quantification of the Vtg standard used in the FHM Vtg ELISA. Important issues here are • the choice of source for purification • the purification method • the quality assurance • the quantification method • stabilization procedure Ideally, a certified reference material (CRM) should be used as a standard in quantitative assays. In the case of a Vtg ELISA, this CRM should consist of intact Vtg purified to apparent homogeneity from the test species in question, and should be quantified according to accepted methods for protein quantification. In addition, the Vtg should be stabilized for shipping and storage. Page 25 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA Due to the lack of guidelines or criteria for these issues, we will not refer to the FHM Vtg standard as CRM, but rather only as Reference Material (RM). *** SOURCE FOR RM PURIFICATION Various sources of purified standards for Vtg assays have been used, including plasma, WBH, liver homogenate, ascites fluid, and egg yolk. For each of these sample types, the state of the Vtg will be different. For example, the liver cell will contain immature (unprocessed) Vtg that has not undergone full post-translational modifications, as well as mature Vtg ready for secretion, whereas the egg yolk will contain the lipovitellin form processed after uptake. WBH will contain a mixture of these (unless ovaries and /or liver have been removed), in addition to a high level of proteolytic activity that may act to degrade the protein during preparation. We chose to purify Vtg from plasma obtained from estrogenized FHM, because such plasma contains intact, circulating Vtg at a high concentration (mg/ml levels). Plasma was kindly provided by Charles Tyler, University of Exeter, UK. The matrix for RM production should be obtained from estrogenized fish exposed to 17ß-oestradiol (or another given reference oestrogen) for a given period of time. A suitable protease inhibitor (e.g. aprotinin or a protease inhibitor cocktail) should be added to the matrix during sampling to avoid degradation of Vtg. *** PURIFICATION OF RM Due to the instability of Vtg, the purification procedure should be as rapid as possible while maintaining the integrity of the protein and yielding a pure product. Various methods include • • • Ion exchange chromatography, with or without a following gel permeation clean-up (e.g. Brion et al., 2002). Selective precipitation of Vtg from plasma using MgCl2. This is a rapid method that has been successfully used with some species (e.g. trout and carp: Norberg & Haux, 1985, and Nilsen et al., 2003), but appears to be less useful for other species. Immunoaffinity-based procedures (e.g. chromatography or magnetic beads). This strategy requires Vtg-specific Abs, and may give a bias in the composition of the purified product depending on the epitope specificity of the antibody used. Different purification protocols may lead to different compositions of the Vtg holoprotein (i.e. different parts of the phospholipoglyco-modifications may be retained), which also may lead to different affinities of the antibodies in an ELISA. Using either of these methods, mg quantities of purified Vtg can be obtained within a short period of time (10-30 min). To prevent degradation of the protein this process Page 26 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA should be performed in a cold environment using cold buffers containing protease inhibitors. We chose to use anion-exchange chromatography to purify the RM for the FHM Vtg ELISA. Using spin columns from Vivascience (Hannover, Germany), Vtg was rapidly purified in sufficient quantities from small amounts of plasma from estrogenized FHM. *** QUALITY ASSURANCE OF RM The purity and homogeneity of the RM needs to be established using gel electrophoresis. Both SDS-PAGE and 2-dimensional electrophoresis (2-DE) were carried out to demonstrate the purity of the RM (Figure 4). In addition, Matrix assisted laser desorption ionization mass spectrophotometry (MALDI-MS) was performed to obtain more information about the proteins observed on the 2-DE gel. Both SDS-PAGE and 2-DE shows few impurities. In SDS-PAGE, three main bands are clearly visible. All three bands are recognised by the FHM Vtg-specific mAb 1E9 in western blot. Peptide sequencing of the spots “FHM1” and “FHM2”, obtained by MALDI-MS, were both recognised as FHM Vtg using the Mascot database (http://www.matrixscience.com). “FHM1” had hits in different parts of the protein, whereas “FHM2” only had hits in the N-terminal part, indicating that it is a breakdown product of the main 150 kDa Vtg holoprotein. “FHM3” was not recognised as Vtg, but must be seen as an unknown, low-molecular weight peptide contaminant. All in all, the results show that the purified protein is fathead minnow Vtg, and that the preparation is satisfactory pure. Page 27 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA a b 1 kDa 2 3 4 5 6 7 250 150 100 75 50 37 c kDa FHM1 FHM2 97.4 66.2 FHM3 Figure 4 a: SDS-PAGE with different amounts of FHM Vtg. 7% separation gel. Lane 1: Broad range standard (Bio-Rad). Lane 2: Empty. Lane 3: 1 µg Vtg. Lane 4: 2 µg Vtg. Lane 5: 3 µg Vtg. Lane 6: Empty. Lane 7: 5 µg Vtg. Figure 4 b: Western blot. 1 µg Vtg was applied, and mAb 1E9 was used to detect the FHM Vtg Figure 4 c: 2-DE of FHM Vtg. 50 µg Vtg was applied. pH 3-10, 9% separation gel, low range standard (Bio-Rad). The indicated proteins (FHM 1-3) were analysed with MALDI-MS. *** QUANTIFICATION OF RM Various methods are commonly used to quantify purified Vtg. All are dependent on a pure product. • Staining methods such as Lowry or Bradford rely on the use of a protein standard, normally bovine serum albumin (BSA), and assume a similar staining response of the protein in question to this standard protein. In many cases, this may not be true; especially for a protein like Vtg, which contains various non-peptide groups (sugars, phosphates, lipids). Page 28 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA • • Measurement of absorbance at 280 nm is a simple and commonly used quantification method. The method relies on the use of an extinction coefficient, which is different for each protein, due to the intramolecular environment affecting the exposure of UV absorbing aromatic amino acid residues. A theoretical extinction coefficient can be calculated from the full amino acid sequence of a protein, however posttranslational modifications will affect this value. A more precise method of protein quantification is quantitative amino acid analysis. This analysis can be performed by independent analytical laboratories using standard instrumentation. The amino acid composition can be compared to the theoretical composition if the sequence is known. A limitation of this method is that only the protein portion of the Vtg molecule is quantified. The lipid and phosphate parts have been reported for some species to represent 15–20% and 0.6–0.8%, respectively (e.g. Silversand & Haux, 1995), whereas the carbohydrate portion is not well studied. We feel that the reliability and independence of amino acid analysis outweighs the limitations, and we chose this method for quantification of the FHM Vtg RM. Two parallel analyses were performed at the Peptide Synthesis Lab at the Biotechnology Centre of Oslo, Norway. *** STABILIZATION OF RM Vtg is sensitive to freeze-thaw cycles (Figure 5), and needs to be stored at –80°C and shipped on dry ice. In the Biosense laboratory, we have developed a lyophilisation procedure that gives good stability to purified Vtg, with more than one year’s stability of Vtg at 4°C (these stability data are “real time” data, and are so far based on other species than FHM). After lyophilisation, the RM Vtg was calibrated against non-lyophilised Vtg in the FHM ELISA. FHM Vtg freeze/thaw cycles % of theoretical amount 100 80 60 40 20 0 0 1 2 3 4 5 Numer of freeze/thaw cycles Figure 5: Stability of lyophilised FHM Vtg during repeated freeze-thaw cycles. One vial of reconstituted FHM Vtg was exposed to repeated freezing and thawing. The amount of Vtg in the vial was analysed in the FHM Vtg ELISA. Page 29 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA The FHM Vtg RM was compared to FHM Vtg supplied as a standard for the US EPA method study (http://www.epa.gov/scipoly/oscpendo/assayvalidation/status.htm). This Vtg had been purified using anion-exchange chromatography and had also been quantified using amino acid analyses, but the Vtg solution had been thawed once before this comparison was made. Figure 6 shows that there are between 1.2 and 1.6-fold difference between the absorbance values obtained with the two standards. This small difference is most likely due to the state of degradation and purity of the Vtg. FHM standards 10 y = 0.2018x0.9864 A450-NSB R2 = 0.9992 y = 0.1514x0.9523 1 R2 = 0.9988 Biosense Standard 0.1 US EPA Standard Power (Biosense Standard) Power (US EPA Standard) 0.01 0.01 0.1 1 10 100 ng/ml Figure 6: Comparison of the FHM Vtg RM with FHM Vtg supplied by the US EPA for Vtg assay comparison study. The standards were analysed in the FHM Vtg ELISA. *** SAMPLE PREPARATION AND SPIKING Sample blanks: Plasma from male fathead minnows was purchased from Fish Soup, Newberry, Florida, USA. Plasma from 34 individual males was screened, and samples having non-detectable levels of Vtg at a 1:100 dilution were pooled. WBH from male fathead minnows was a gift from Robert Bringolf, Iowa State University, USA. The WBH showed no detectable levels of Vtg at a 1:100 dilution. Spiked samples: For spiking/recovery studies, Sample blanks (plasma, WBH and kit Dilution buffer) were diluted 10-fold with kit Dilution Buffer containing different concentrations of purified, non-lyophilised Vtg. Samples were mixed thoroughly, aliquoted into suitable volumes and frozen at –80oC. Concentrations in spiked samples were 5, 25 and 125 ng/ml. Naturally incurred samples: Plasma with low levels of Vtg was purchased from Fish Soup, Newberry, Florida, USA. WBH with low levels of Vtg was a gift from Robert Bringolf, Iowa State University, USA. *** Page 30 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA VALIDATION RESULTS APPLICABILITY Applicability (Scope): The analytes, matrices and concentrations for which a method may be used satisfactorily. This validation was performed in order to determine the characteristics of the FHM Vtg ELISA, developed for analyses of Vtg in the fathead minnow (Pimephales promelas), as an endpoint in endocrine disrupting chemicals (EDC) screening tests. The ability of the ELISA to accurately analyse Vtg levels in spiked (fortified) plasma, whole body homogenate (WBH) and the ELISA kit Dilution buffer was analysed. Four levels of Vtg were used in this validation (0, 0.5, 2.5 and 12.5 ng/ml Vtg), corresponding to different levels within the standard curve working range (0.1-25 ng/ml). *** CALIBRATION Calibration, is the empirical determination of the relationship between the parameter measured (e.g. ELISA absorbance) and the analyte (Vtg) concentration. The range of concentrations of analyte where such relationship is established is often referred to as "calibration range" or the "standard curve working range". Calibration of the assay was performed with purified, lyophilised FHM Vtg RM. A standard serial dilution containing 11 concentration points was used, and a log-log curve fit (using Microsoft Excel) was used to define the relationship between concentration and response (absorbance). Standard curves from 15 assays were compiled (Figure 6, Table 3 a-b). Three standard curves were run each day for five days. The standard serial dilutions were prepared fresh each day, and the same solution was used on three different ELISA plates. The Within-Day and Between-Day Precision was calculated (Figure 6). FHM Vtg standard 100 10 90 80 70 60 50 0.1 40 %CV A450-NSB 1 30 0.01 20 10 0.001 0.01 0 0.1 1 10 100 ng/ml Standard Within-Day Precision Between-Day Precision Figure 6: Combined FHM Vtg standard curve, average from 15 assays (using Microsoft Excel). The secondary y-axis shows the Within-Day and Between-Day Repeatability Precisions (RSDr) for the standard curves. Page 31 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA Table 3a: Within-Day Repeatability Precision for FHM Vtg standard curves Within Day Precision Concentration (ng/ml) NSB 0.05 0.10 0.20 0.39 0.78 1.56 3.13 6.25 12.50 25.00 50.00 Day 1 Average absorbance (450 nm), 3 standard curves %CV 0.061 0.069 0.072 0.084 0.108 0.167 0.276 0.507 0.918 1.822 3.338 5.307 8.2 11.1 8.7 5.3 4.2 4.4 4.0 4.5 4.4 2.2 2.8 16.1 Average absorbance (450 nm), 3 standard curves %CV 0.060 0.065 0.072 0.083 0.110 0.166 0.278 0.506 0.940 1.811 3.194 5.793 9.0 5.9 5.3 6.2 8.7 10.2 10.1 11.5 11.0 10.0 8.7 7.3 Average absorbance (450 nm), 3 standard curves %CV 0.057 0.062 0.069 0.079 0.102 0.153 0.255 0.460 0.849 1.657 3.027 5.263 5.3 3.3 5.7 3.6 3.2 4.6 6.5 6.6 5.7 5.8 5.2 13.9 Average absorbance (450 nm), 3 standard curves %CV 0.055 0.060 0.066 0.074 0.094 0.140 0.215 0.416 0.735 1.444 2.657 4.358 5.6 5.0 5.9 2.3 3.7 5.8 5.1 6.3 5.2 5.1 4.2 12.4 Average absorbance (450 nm), 3 standard curves %CV 0.056 0.063 0.069 0.082 0.112 0.170 0.271 0.498 0.945 1.661 3.009 5.017 4.1 2.1 1.6 4.8 4.4 6.4 8.4 9.4 7.6 11.5 3.8 17.4 Overall Within-Day Precision 6.4 5.5 5.4 4.4 4.8 6.3 6.8 7.7 6.8 6.9 4.9 13.4 Overall Within-Day Precision (working range, 0.1-25 ng/ml) 6.0 Day 2 Day 3 Day 4 Day 5 Table 3b: Between-Day Repeatability Precision for FHM Vtg standard curves Between-day Precision Concentration (ng/ml) NSB 0.05 0.10 0.20 0.39 0.78 1.56 3.13 6.25 12.50 25.00 50.00 Average absorbance (450 nm), 5 days 0.058 0.064 0.070 0.080 0.105 0.159 0.259 0.477 0.877 1.679 3.045 5.148 %CV 4.2 5.1 3.8 5.1 6.8 7.8 10.0 8.3 10.1 9.1 8.4 10.2 Overall Between-Day Precision (working range, 0.1-25 ng/ml) 7.7 The results show that standard curves obtained on both the same day and on different days show little variability, with an average Within-Day Precision of 6.0 % and a Between-Day Precision of 7.7 %. The standard curve working range was between 0.1 and 25 ng/ml using a log-log transformation of the data. *** LOD AND LOQ (SENSITIVITY) Limit of Detection is the smallest amount or concentration that can be reliably distinguished from zero. Defined here as reagent blank + 3x standard deviation of reagent blank. Indicates that the analyte is present, but not necessarily allowing exact quantification. Limit of Quantification A concentration above which the analytical method can operate with an acceptable precision. Defined here as reagent blank + 10x standard deviation of reagent blank. Page 32 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA Sample LoQ. The LoQ corrected for minimum dilution factor necessary to avoid matrix effect. Limit of Detection (LoD) and Limit of Quantification (LoQ) were determined from matrix blanks analysed during Precision studies. Since the samples analysed at a 1:100 dilution did not have detectable levels of Vtg, LoD and LoQ were determined from NSB-corrected absorbance levels and “translated” into concentration using the relevant standard curve equation (Table 4a-c). Table 4a) LoD and LoQ for Plasma Run1 Replicate 1 A450nm 0.000 Replicate 2 A450nm -0.003 Replicate 3 A450nm -0.002 Average A450nm -0.001 Stdev A450nm 0.002 3x Stdev A450nm 0.003 LoD ng/ml 0.02 10x Stdev LoQ A450nm ng/ml 0.014 0.11 Run2 0.001 -0.002 -0.003 -0.001 0.002 0.005 0.03 0.019 0.12 Run3 -0.001 -0.001 -0.001 -0.001 0.000 0.000 0.00 0.003 0.02 Run4 0.001 0.002 0.000 0.001 0.001 0.005 0.04 0.013 0.12 Run5 0.000 0.000 -0.003 -0.001 0.002 0.004 0.02 0.017 0.10 0.003 0.02 0.013 0.09 Average Table 4b) LoD and LoQ for WBH Run1 Replicate 1 A450nm -0.004 Replicate 2 A450nm 0.000 Replicate 3 A450nm -0.001 Average A450nm -0.002 Stdev A450nm 0.002 3x Stdev A450nm 0.005 LoD ng/ml 0.04 10x Stdev LoQ A450nm ng/ml 0.020 0.15 Run2 -0.005 -0.006 -0.007 -0.006 0.001 -0.004 ND 0.002 0.01 Run3 -0.002 -0.004 -0.003 -0.003 0.001 -0.001 ND 0.005 0.04 Run4 0.002 -0.003 -0.005 -0.002 0.004 0.009 0.08 0.034 0.32 Run5 -0.002 -0.004 -0.003 -0.003 0.001 0.000 0.00 0.008 0.05 0.002 0.04 0.014 0.11 Average Table 4c) LoD and LoQ for Dilution buffer Run1 Replicate 1 A450nm -0.002 Replicate 2 A450nm 0.001 Replicate 3 A450nm -0.006 Average A450nm -0.002 Stdev A450nm 0.004 3x Stdev A450nm 0.009 LoD ng/ml 0.06 10x Stdev LoQ A450nm ng/ml 0.033 0.24 Run2 -0.009 -0.008 -0.010 -0.009 0.001 -0.005 ND 0.003 0.02 Run3 0.000 0.001 -0.001 0.000 0.001 0.003 0.02 0.009 0.07 Run4 -0.001 -0.001 -0.001 -0.001 0.000 0.000 0.00 0.003 0.03 Run5 -0.002 -0.003 0.001 -0.001 0.002 0.005 0.03 0.019 0.11 0.002 0.03 0.013 0.09 Average The LoD varied between 0.02 and 0.04 ng/ml for different sample types, and the LoQ varied between 0.09 and 0.11 ng/ml. Thus, the LoQ is in good agreement with the lower limit of the standard curve working range (0.10 ng/ml). *** SELECTIVITY (MATRIX EFFECT) In order to determine the degree of interference from sample matrices on quantification of Vtg, plasma and WBH were investigated for adverse effects on the signal response, i.e. plasma or matrix effect. Different dilutions of matrix blanks were Page 33 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA spiked with a range of Vtg concentrations, and recovery was measured and compared. Figure 8 and 9 show that there is an inhibition of the signal at low dilutions of both plasma and WBH, resulting in an underestimation of Vtg at these dilutions. This effect varies somewhat between different samples, and with the spike concentration. Note that some haemolysis had happened during preparation of the plasma, which may have an effect on matrix effect. Based on these results showed in Figure 8 and 9, we recommend 1:50 for plasma and 1:100 for WBH as the minimum dilution factors. Lower dilution factors might be used, but this should be tested in individual laboratories, and may depend on the method of sample preparartion. Plasma Wbh b ) 2.5 2.0 1.5 A450-NSB A450-NSB a)2.0 1.5 1.0 1.0 0.5 0.5 0.0 0.0 50 100 150 200 250 300 350 400 450 500 550 50 Dilution factor (x-fold) 0 0.5 2.5 100 150 200 250 300 350 400 450 500 550 Dilution factor (x-fold) 0 0.5 2.5 12.5 12.5 Parallelism between standard and unspiked samples c) 10 A450-NSB 1 Std Plasma 0.1 Wbh Wbh Power (Plasma) Power (Std) 0.01 Power (Wbh) 0.001 10 100 1000 10000 100000 (Relative) dilution factor (x-fold) Figure 8: Effect of plasma and WBH sample dilution on detection of Vtg. Experiments were performed with spiked samples (Figure 8a-b) or with naturally incurred samples (Figure 8c), containing Vtg. Page 34 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA a) b Plasma Wbh a) ) 100 ng/ml (measured ng/ml (measured 100 10 1 0.1 0.01 0.01 0.1 1 10 10 1 0.1 0.01 0.01 100 0.1 PLASMA 1:50 c) PLASMA 1:100 PLASMA 1:200 d ) 100 Recovery vs dilution Averagwe recovery (%) 120 100 80 Plasma Wbh 60 40 20 STANDARD + 5 µl Average recovery (%) STANDARD 1 10 100 ng/ml (theoretical) ng/ml (theoretical) 0 WBH 1:50 WBH 1:100 WBH 1:200 Plasma 80 60 Plasma 40 20 0 50 100 200 25 Dilution factor 50 100 Dilution factor Figure 9: Effect of plasma and WBH sample dilution on quantification of Vtg. Different amounts of plasma or WBH were added to standard dilutions (Figure 9a-b). The average recovery for each sample dilution factor was cacluated (Figure 9c). An additional experiment was conducted with even lower dilution of plasma, tested with three different Vtg levels (Figure 9d). *** REPEATABILITY PRECISION (INTRA- AND INTERASSAY VARIATION), RSDr Precision is the closeness of agreement between test results obtained under stipulated conditions. • Repeatability Precision (same laboratory and operator, samples, equipment, short time intervals), separated into Within-Day and Between-Day Repeatability precision, usually expressed as relative standard deviation, RSDr. Matrix blanks from plasma and WBH, as well as ELISA kit Dilution buffer were analysed, spiked with three different concentrations of Vtg corresponding to the low, medium and high parts of the standard curve working range. In addition, unspiked material was analysed. All samples had been aliquoted and stored at –80oC until analysis. On five successive days, three aliquots were thawed and analysed in triplicates at a 1:100 final dilution. Within-Day and Between-Day Repeatability Precision (RSDr), often referred to as intra- and interassay variation, were calculated (Table 5a-c and Table 6a-c). Within-Day Repeatability Precision: %CV between the three aliquots analysed in one day. Page 35 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA Overall Within-Day Repeatability Precision: average of the individual Precision values for five days Table 5a) Within-Day Precision for spiked Plasma samples Run # 1 2 3 4 5 Spike concentration ng/ml 0.5 Replicate 1 ng/ml 0.3 Replicate 2 ng/ml 0.3 Replicate 3 ng/ml 0.3 Average ng/ml 0.3 RSDr % 0.7 2.5 1.9 1.8 1.9 1.9 3.0 12.5 10.8 10.5 10.8 10.7 1.5 0.5 0.3 0.3 0.3 0.3 2.6 2.5 1.8 1.7 1.7 1.7 3.4 12.5 9.9 9.4 9.2 9.5 3.7 0.5 0.4 0.3 0.3 0.3 8.8 2.5 2.0 1.9 1.9 1.9 3.7 12.5 10.9 10.3 10.7 10.6 2.8 0.5 0.4 0.4 0.3 0.4 5.8 2.5 2.1 1.9 1.9 2.0 4.0 12.5 12.6 11.3 11.8 11.9 5.3 0.5 0.3 0.3 0.3 0.3 7.8 2.5 1.6 1.7 1.6 1.6 3.1 12.5 9.9 10.0 9.8 9.9 0.6 Overall Within-Day Precision (%) 3.8 Table 5b) Within-Day Precision for spiked WBH samples Run # 1 2 3 4 5 Spike concentration ng/ml 0.5 Replicate 1 ng/ml 0.3 Replicate 2 ng/ml 0.3 Replicate 3 ng/ml 0.3 Average ng/ml 0.3 RSDr % 4.6 2.5 1.6 1.5 1.5 1.6 4.1 12.5 8.0 7.2 7.4 7.5 5.5 0.5 0.3 0.3 0.3 0.3 4.7 2.5 1.5 1.5 1.5 1.5 3.3 12.5 7.9 7.4 7.5 7.6 3.4 0.5 0.3 0.3 0.3 0.3 4.4 2.5 1.8 1.9 1.8 1.8 2.4 12.5 9.1 9.5 9.5 9.4 2.6 0.5 0.4 0.3 0.3 0.4 7.0 2.5 2.0 1.7 1.8 1.8 7.7 12.5 10.3 9.0 9.6 9.7 6.5 0.5 0.3 0.3 0.3 0.3 10.1 2.5 1.9 1.7 1.7 1.8 7.3 12.5 9.6 9.1 8.8 9.2 4.3 Overall Within-Day Precision (%) 4.6 Table 5c) Within-Day Precision for spiked Dilution buffer samples Page 36 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA Run # 1 2 3 4 5 Spike concentration ng/ml 0.5 Replicate 1 ng/ml 0.5 Replicate 2 ng/ml 0.4 Replicate 3 ng/ml 0.4 Average ng/ml 0.4 RSDr % 10.3 2.5 2.1 2.2 2.1 2.1 2.3 12.5 11.1 11.1 11.3 11.1 0.9 0.5 0.3 0.3 0.3 0.3 3.7 2.5 1.8 1.8 1.9 1.9 2.5 12.5 11.2 10.6 10.3 10.7 4.2 0.5 0.4 0.4 0.4 0.4 3.1 2.5 2.2 2.2 2.2 2.2 1.2 12.5 11.4 11.1 11.4 11.3 1.9 0.5 0.4 0.4 0.4 0.4 2.4 2.5 2.3 2.3 2.4 2.3 2.3 12.5 11.6 11.7 12.2 11.8 2.6 0.5 0.4 0.4 0.3 0.4 4.2 2.5 2.2 2.1 1.5 2.0 20.8 12.5 12.3 11.4 11.0 11.6 5.9 Overall Within-Day Precision (%) 4.6 Overall Within-Day Precision for all sample types and concentrations 4.5 Between-Day Repeatability Precision: % CV between the average measured concentrations from each day. Overall Between-Day Repeatability Precision: Average Precision for all concentrations Table 6a) Between-Day Precision for Plasma samples Spike concentration ng/ml) 0.5 Average (ng/ml) 0.3 RSD % 11.5 2.5 1.8 8.2 12.5 10.3 8.8 r Overall Between-Day Precision 9.5 Table 6b) Between-Day Precision for WBH samples Spike concentration ng/ml) 0.5 Average (ng/ml) 1.8 RSD % 1.9 2.5 1.6 10.3 12.5 5.8 17.4 r Overall Between-Day Precision 9.9 Page 37 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA Table 6c) Between-Day Precision for Dilution buffer samples Spike concentration ng/ml) 0.5 Average (ng/ml) 0.4 RSD % 14.7 2.5 2.1 11.4 12.5 11.1 4.7 r Oerall Between-Day Precision 10.3 Overall Between-Day Precision for all sample types and concentrations 9.9 The results show low variation in quantification, both within the same day (Overall Within-Day Precision = 4.5 %) and between successive days (Overall Between-Day Precision = 9.9 %). *** ACCURACY Accuracy is the closeness of agreement between a test result and the accepted reference value of the property being measured. Recovery is the proportion of the amount of analyte, present in or added to, the analytical portion, which is extracted and presented for measurement Bias is the difference between the test results and an accepted reference value Recovery and bias The concentrations measured in the spiked samples during Precision studies were compared to the theoretical values and Recovery and Bias were determined using the following formulas (Table 7-8): Recovery = (C1-C2)/C3 x 100 Where C1= concentration measured in spiked sample, C2= concentration measured in unspiked sample, C3= theoretical concentration. Bias = (C3-(C1-C2))/C3 x 100 Where C1= concentration measured in spiked sample, C2= concentration measured in unspiked sample, C3= theoretical concentration Table 7: Recovery (expressed as % of theoretical concentration) Spike concentration (ng/ml) Plasma WBH Spiked buffer 0.5 64.2 63.3 75.9 2.5 72.8 68.0 83.9 12.5 84.3 69.4 90.5 Overall Recovery (%) 73.8 66.9 83.4 Overall Recovery for all sample types and concentrations (%) Page 38 74.7 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA Table 8: Bias (expressed as % difference from theoretical value) Spike concentration (ng/ml) Plasma WBH Spiked buffer 0.5 -35.8 -36.7 -24.1 2.5 -27.2 -32.0 -16.1 12.5 -15.7 -30.6 -9.5 Overall Bias (%) -26.3 -33.1 -16.6 Overall Bias for all sample types and concentrations (%) -25.3 The results show that the recovery and bias depend on both sample type and spike concentration, with an overall 25% underestimation of spike concentrations in the sample. Using freshly spiked samples, without previous freezing/thawing of the samples, Recovery was measured during the Selectivity study (see Figure 9). Results from these experiments, using several different concentrations of Vtg (standard curve dilution series), and different dilutions of plasma and WBH, gave the Recovery results much closer to 100% (Table 9a-c). Table 9a: Recovery in WBH and plasma: Average Recovery for 11 different spike concentrations (expressed as % of theoretical value) Dilution factor Plasma WBH 1:50 81.4 102.0 1:100 106.7 102.0 1:200 97.6 105.9 Table 9b: Recovery in WBH and plasma: Average Recovery for three different spike concentrations (expressed as % of theoretical value) Dilution factor Plasma WBH 1:25 65.6 nd 1:50 75.9 84.8 1:100 95.2 102.5 Table 9c: Recovery in WBH and plasma, average of two experiments (see Table 9a and b) Dilution factor Plasma WBH 1:25 65.6 nd 1:50 78.7 93.4 1:100 101.0 102.3 1:200 97.6 105.9 *** RUGGEDNESS Ruggedness. The ability of the measurement process to resist changes in results when subjected to minor changes in environmental and procedural variables. In order to investigate the stability of the assay when exposed to variations in the environment and assay procedure, seven parameters were combined in eight assays to determine their effect on quantification in the FHM Vtg ELISA (according to Youden & Steiner, 1975). The effect on quantification of the three matrix blanks, spiked with three different concentrations, was analysed. Table 10 shows the compiled results. Page 39 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA Table 10: Ruggedness test. Standard conditions are shown in bold. Alterations higher than 10% are highlighted. Condition altered Value of condition Sample type Sample concentration level low med Difference (%) Buffer temperature RT2 30 C Incubation temperature o Standard/sample incubation time Detection Ab incubation time 4 6 -10 -13 -11 WBH 4 -9 -12 -5 Buffer 12 11 4 9 Plasma -18 -18 -21 -19 WBH -7 -5 -8 -7 Buffer -2 1 7 2 Plasma 6 7 11 8 WBH 1 4 8 4 Buffer 5 4 -3 2 Plasma -1 -10 -11 -7 WBH 3 1 -5 0 Buffer 8 3 -6 2 Plasma 16 11 -1 9 WBH 11 7 -5 4 Buffer 7 0 1 3 Plasma 9 8 4 7 WBH 4 3 1 3 Buffer -8 -5 1 -4 Plasma -2 1 6 2 WBH -7 -4 0 -3 RT 4C o Development time 6 -11 5 3 TMB solution temperature 7 Plasma 0.5 h 1h before Buffer 1.5 h 1h washes Average (%) 1 Cold RT Number of development high 1 20 min 30 min 1 Minus sign denotes that the value for the unaltered condition was lower than for the altered condition 2 RT in this assay was 23oC The results show that buffer and incubation temperature have the highest influence on Vtg quantification in the FHM Vtg ELISA. The effect of buffer temperature was studied in separate assays, and showed that room-tempered buffer had a stronger effect on the samples than on the standard, increasing absorbance and the measured Vtg concentration in the samples (data not shown). These results demonstrate the importance of keeping the Dilution buffer cold and to keep the room temperature relatively low (20-25oC). On the other hand, deviations to the incubation times have less influence on the quantification of samples. *** Page 40 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA COMPARISON WITH EXISTING METHODS No established reference method for measuring FHM Vtg exists today, so the new FHM Vtg ELISA was therefore compared with two other commercially available ELISA kits, a Carp Vtg ELISA (Biosense Laboratories) and a FHM Vtg ELISA (competitor). *** Carp Vtg ELISA (Biosense Laboratories AS) Plasma and WBH samples from the US EPA FHM Vtg assay comparison study (2003) were tested in the FHM Vtg ELISA. These samples had previously been tested in the Biosense Carp Vtg ELISA, using both the kit (carp) Vtg standard and the FHM Vtg standard supplied with the study samples. Figure 10a and 10d show the results compared. The Carp Vtg ELISA utilises one monoclonal and one polyclonal carp Vtg-specific antibodies, as well as carp Vtg standard, and the assay is therefore heterologous for the FHM. However, the antibodies show excellent cross-reactivity with FHM Vtg (Nilsen et al 2004), reflected in values varying less than 2-fold between the two assays and a good correlation between Vtg levels measured in the samples (R2 > 0.99). *** Competitor FHM ELISA kit The Biosense and a competitor FHM Vtg ELISA kit were compared using spiked and unspiked samples with different Vtg levels. Figure 10b, c and e show the two standard curves and the results from the analysed samples. Although the absolute values varies on average less than 2-fold, due to differences in standards and antibodies, the results show very good correlation (R2 > 0.99). The largest difference is the sensitivity of the assays, with a 78 times more sensitive standard curve in the Biosense FHM ELISA, and a corresponding lack of detection of Vtg in samples spiked with low Vtg concentrations in the competitor ELISA kit. *** Page 41 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA Plasma samples a) 1000000 µg/ml in plasma 100000 10000 1000 100 10 1 0.1 A B E G FHM ELISA 4488 463.9 303.3 6943 C ELISA, C standard 6719 169.0 0.326 522.5 D 6388 R S 0.255 4418 49773 48807 52006 507.4 25156 0.252 0.209 0.253 0.310 20073 30526 0.259 H I K L M N P 6727 67459 85811 58270 318.8 0.427 0.272 0.222 0.399 T U W Y C ELISA, FHM standard 10893 344.8 0.552 740.7 10385 34537 0.434 0.371 0.443 0.525 27518 41963 0.450 10904 95532 12148 82559 631.6 Sample ID b) Standard curves 10 y = 0.1443x1.0271 R2 = 0.9986 A450-NSB 1 Biosense Competitor 0.1 Power (Biosense) 0.01 Power (Competitor) y = 0.0039x1.0359 0.001 0.01 0.1 1 10 100 R2 = 0.9986 1000 ng/ml Vtg Biosense FHM Vtg ELISA vs competitor FHM ELISA c) 10000000 1000000 100000 ng/ml 10000 1000 Biosense 100 Competitor 10 1 0.1 PJ Biosense PQ PB PS 0.36 2.12 11.4 WJ Competitor WB 1.54 WS 8.66 BJ BQ BB BS WM WA WR 0.35 2.16 11.4 6.89 25.2 45011 996 38334 46661 84219 1956 84441 74427 14.63 100000 d) WQ 0.30 21.5 36.9 PA PR e) 2 R = 0.9937 10000000 10000 2 R = 0.9911 1000000 100000 Wbh samples Plasma samples 100 Power (Wbh samples) Power (Plasma samples) 10 Competitor kit 1000 Carp kit PM 10000 R2 = 0.9988 1000 100 10 1 1 0.1 0.1 0.1 0.1 1 10 100 1000 10000 1 10 100 1000 10000 100000 1E+06 1E+07 Biosense kit 100000 FHM kit Figure 9: Comparison of the Biosense FHM Vtg kit with two methods. Figure 9a: Samples from a US EPA method comparison study were compared in the Biosense Carp (C) and FHM Vtg ELISA kits, using both C and FHM Vtg standard in the C ELISA. Figure 9b: FHM Vtg kit standard curves in the Biosense and competitor FHM Vtg ELISA kit. Figure 9c: Identical samples analysed in the Biosense and competitor FHM Vtg ELISA kit. 9d: Correlation between results obtained with Biosense Carp and FHM Vtg ELISA kits. 9e: Correlation between Biosense and competitor FHM Vtg ELISA kits. *** Page 42 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA DISCUSSION A new sandwich ELISA, based on monoclonal antibodies raised against FHM Vtg, has been developed to quantify Vtg in samples from FHM. Vtg was purified from FHM plasma to be used as standard in the ELISA,. The Vtg was further characterised and quantified, and was found to be of sufficient purity to be used reliably as Reference Material (RM) in the FHM ELISA. A Single-Laboratory Validation was performed to validate the performance of the new Biosense FHM Vtg ELISA. A set of plasma, WBH and ELISA kit Dilution buffer samples, both spiked, unspiked and naturally incurred (containing Vtg), were analysed on different days. Variation within days and between days was determined, as well as the ability of the assay to measure correct values in spiked samples. A calibration curve was run, prepared from a serial dilution of the FHM Vtg RM, in each assay. Using log-log transformation of the standard curves, the working range was from 0.1-25 ng/ml (250-fold). For 15 standard curves (three separate standard curves per day for five days), the Within-Run and Between-Run Repeatability Precision (RSDr) within the working range was 6.0 and 7.7%, respectively. This shows that the variability between different vials of Vtg RM is low, as is the variability between standard curves prepared on different days. Also, the 250-fold working range is well within our aim (100-fold, Goksøyr et al, 2003) Vtg concentrations in samples can vary over several orders of magnitude, and a broad working range is important because fewer dilutions from each sample of unknown Vtg concentration are necessary in order to “hit” the standard curve working range. Important parameteres for a quantitative assay is the Limit of Detection (LoD) and Limit of Quantification (LoQ). The FHM assay is highly sensitive, with an average LoD of 0.03 ng/ml and an LoQ equal to the lower limit of the standard curve working range, 0.10 ng/ml. With minimum recommended sample dilutions of 1:50 for plasma and 1:100 for WBH, the sample LOQs were 4.7 and 11.4 ng/ml, respectively. Both plasma and WBH from unexposed male fish (from the US EPA Study) were easily quantified by the FHM ELISA. In order to determine the Within-Day and Between-Day Repeatability Precision (RSDr) of the ELISA, three replicates of each sample were analysed on five different days. Within-Day RSDr values were between 3.8 and 4.6 % for the different sample types, with an overall RSDr of 4.5%. The Between-Day RSDr was between 9.5 and 10.3%, with an overall RSDr of 9.9%. These values show that the assay shows little variability between runs, and are well within the aims defined by Goksøyr et al (2003). The assay’s ability to accurately quantify Vtg in spiked samples was analysed by comparing concentrations measured in the ELISA with the theoretical concentrations. Samples with three different concentrations of Vtg were analysed, Page 43 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA and the Recovery varied between 63 and 91%, depending on concentration and sample type, with an overall Recovery of 75% (Bias = -25%). Vtg is an unstable molecule, prone to degradation (Arukwe and Goksøyr 2003). For this reason, spiking of samples should ideally be done on the day of analysis to avoid freeze-thaw cycles and degradation. However, in order to obtain data for determination of Between-Day Repeatability Precision, samples had to be diluted, aliquoted and frozen. This is likely to have affected the Recovery of Vtg in spiked samples. Spiking/recovery experiments performed without freezing and thawing of the spiked samples, gave Recovery in both plasma and WBH between 79-106%, supporting this theory. The Ruggedness test revealed that small changes to factors such as incubation time and temperatures affected the FHM ELISA’s ability to quantify Vtg to different degrees. Seven factors were modified and tested together in different combinations according to Youden & Steiner (1975), and the factors having highest impact on Vtg quantification were elevated buffer and room temperatures. Since no official Reference Method for FHM Vtg quantification is established, the new Biosense FHM ELISA was compared to two exisiting Vtg ELISA kits. The Biosense Carp Vtg ELISA, although heterologous to the FHM, has successfully been applied on samples from FHM (Nilsen et al, 2004). A set of FHM samples from the US EPA method comparison study (2003) was re-analysed in the FHM ELISA. The absolute values obtained with the two kits differed less than 2-fold, and the correlation (R2) between the results were >0.99, showing that the two kits give comparable results. An alternative FHM ELISA kit was also tested, using a set of spiked and unspiked samples. The Biosense FHM kit standard curve was 78-fold more sensitive than the competitor ELISA Vtg standard, but the absolute values varied less than 2-fold and the correlation between the results from the two kits was equally good, with R2>0.99. The results from these two comparisons show that although absolute values may differ, due to factors such as differences in antibodies, Vtg standard quantification, purity, the assays still yield similar correlating results and suit their purpose of differentiating between control and exposed groups of fish in studies of endocrine disruption. *** CONCLUSION A successful Single-Laboratory Validation was performed on the Biosense FHM Vtg ELISA. The results, summarised in Table 11, show that our pre-defined aims (Goksøyr et al 2003) were met with good margin. The kit is sensitive and reliable, and is therefore a good tool which fits its purpose for quantitative analysis of Vtg in the fathead minnow. Page 44 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA Table 11: Summary of Single-Laboratory Validation results for the Biosense FHM Vtg ELISA kit Performance characteristics Aim1 Value Selectivity Matrix blank < LOD No response at minimum dilution (with the necessary dilution factor Minimum dilution = to avoid matrix effects) 1:50 (plasma), 1:100 (WBH) Calibration Standard curve working range Standard curve working range >10-fold, preferably 50-100 fold 0.1-25 ng/ml (250-fold) to be practical with the dynamic range found in Vtg levels Accuracy (Recovery) Ideally 50-200% 75%2) 79-106%3) 4) Repeatability <20% Within-Day RSDr: 4.5% Between-Day RSDr: 9.9% Limit of Detection (LOD) <10 ng/ml 0.02 ng/ml (plasma) 0.04 ng/ml (WBH) 0.03 ng/ml (buffer) Limit of Quantification (LOQ) <10 ng/ml 0.09 ng/ml (plasma) 0.11 ng/ml (WBH) 0.09 ng/ml (buffer) Sample LOQ 200 – 500 ng/ml 4.68 ng/ml (plasma, 1:50) (=LOQ x necessary matrix dilution) 11.35 ng/ml (WBH, 1:100) 1) Goksøyr et al 2003 2 ) From Precision studies, samples frozen and thawed once, dilution 1:100 3 ) From Selectivity studies, samples freshly spiked and not frozen, dilutions 1:50-1:100 4) Referred to in this document Within-Day and Between-Day Repeatability Precision, RSDr *** REFERENCES AND LINKS AOAC “Harmonisation of analytical terminology in accordance with international standards” http://www.aoac.org/intaffairs/analytical_terminology.htm) Arukwe A, Goksøyr A (1998). Xenobiotics, xenoestrogens and reproduction disturbances in fish. Sarsia 83:225-241. Arukwe A, Goksøyr A (2003). Eggshell and egg yolk proteins in fish: hepatic proteins for the next generation. Oogenetic, population, and evolutionary implications of endocrine disruption. Comp Hepatol 2:4. Brion F, Nilsen BM, Eidem JK, Goksøyr A, Porcher JM (2002). Development and validation of an enzyme-linked immunosorbent assay to measure vitellogenin in the zebrafish (Danio rerio). Environ Toxicol Chem 28: 1699-1708. Crowther JR (2001). The ELISA Guidebook. In: Methods Mol Biol 149. Humana Press, Totowa, NJ Goksøyr A. et al (2003) On the need for a standardized set-up for validation studies of fish vitellogenin assays as an endpoint in endocrine disruptor testing and screening – a proposal. http://www.biosense.com/Docs/GoksoyrEtal2003.pdf Jobling S, Nolan M, Tyler CR, Brighty G, Sumpter JP (1998). Widespread sexual disruption in wild fish. Environ Sci Technol 32: 2498–2506. Page 45 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA Kime DE (1995). The effects of pollution on reproduction in fish. Rev Fish Biol Fisheries 5:52-96 Mommsen TP, Walsh PJ (1988). Vitellogenesis and oocyte assembly. In: Hoar WS, Randall VJ (eds) Fish Physiology XIA. Academic Press, New York, pp 347-406. Nilsen B.M. et al 2004. Development of quantitative vitellogenin-ELISAs for fish test species used in endocrine disruptor screening. Anal Bioanal Chem 378:621 http://www.springerlink.com, DOI 10.1007/s00216-003-2241-2 Norberg B, Haux C (1985). Induction, isolation and a characterization of the lipid content of plasma vitellogenin from two Salmo species: rainbow trout (Salmo gairdneri) and sea trout (Salmo trutta). Comp Biochem Physiol 81B:869–876. Porcher J-M. et al (2003). Intercomparison of zebrafish vitellogenin quantification methods. Society of Environmental Toxicology and Chemsitry, 24th annual meeting North America, http://abstracts.co.allenpress.com/pweb/setac2003/document/?ID=30012 Purdom CE, Hardiman PA, Bye VJ, Eno NC, Tyler CR, Sumpter JP (1994). Estrogenic effects of effluents from sewage treatment works. Chem Ecol 8: 275-285. Silversand C, Haux C (1995). Fatty acid composition of vitellogenin from four teleost species. J Comp Physiol 164B: 593-599. Single Laboratory Validation of Analytical Methods for Dietary Supplements (2003). AOAC International Training Course in Atlanta, Georgia, USA Sumpter JP, Jobling S (1995). Vitellogenesis as a biomarker for estrogenic contamination of the aquatic environment. Environ Health Perspect 103 (Suppl 7): 173-178. The Fitness for Purpose of Analytical Methods (1998). Eurachem, http://www.eurachem.ul.pt/guides/valid.pdf Thompson et al, 2002. Harmonized guidelines for Single-Laboratory validation of methods of analysis. IUPAC Technical Report, Pure Appl Chem 74 (5): 835-855 US EPA Endocrine Disruptor Screening Program, Assay status table. http://www.epa.gov/scipoly/oscpendo/assayvalidation/status.htm Youden & Steiner (1985). In Use of statistics to develop and evaluate analytical methods. AOAC International, Gaithersburg, MD. *** Page 46 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA B: LETTER TO PARTICIPANTS Dear Colleague, Bergen, December 6th, 2004 Validation material – Fathead minnow Vtg ELISA kit and samples We hope that the material arrived safely and in time. I would like you to do the following immediately upon receipt - please note: 1. The state of the samples – samples were shipped on dry ice. There should be dry ice left in the box, and the samples should be frozen. 2. The temperature in the kit parcel – the cooling packs should still be cold. 3. Transfer kit box to a refrigerator at approximately +4-8˚C immediately for storage. 4. Transfer samples to freezer (preferably –80˚C) immediately for storage. 5. Please report the above when returning the Report Sheet (see Study Protocol). 6. Please confirm the arrival of the kits by reporting to janne@biosense.com The shipment contains one custom assembled fathead minnow (FHM) Vtg ELISA kit (V01018402 Pilot kit). For convenience, we have prepared two packs of two ELISA plates each. There are also two vials of Dilution buffer concentrate and TMB substrate, and two vials of Vtg standard. Half of the components are to be used on day one, the other half on day two. Note that only one vial of Detecting antibody (vial E) is included, this must be u sed on both assay days. A Study Protocol is sent as a s eparate file. This contains an o verview of enclosed samples, a study protocol with plate layouts and Report Sheets. Please return Report and Result sheets electronically to janne@biosense.com. Again thank you for your interest to participate in the validation, and good luck with the analysis! All the best Janne K. Eidem Janne K. Eidem Scientist, R&D Biosense Laboratories AS Thormøhlensgate 55 NO-5008 Bergen, Norway Tel: +47 55 54 39 80/66 Mob: +47 91 81 77 81 Fax: +47 55 54 37 71 *** Page 47 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA C: STUDY PROTOCOL AND REPORT SHEET *** Page 48 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA *** Page 49 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA *** Page 50 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA *** Page 51 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA *** Page 52 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA *** Page 53 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA *** Page 54 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA *** Page 55 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA D: REPORT SHEET (EXCEL) Result sheet day 1 Instructions 1. Paste values into each plate (Edit - Paste special - Values) 2. Remove clear outliers (if %CV > 10) 3. Adjust standard curves 4. Use equation from each standard curve to calculate Vtg concentrations in samples Day 1 A B C D E F G H Plate 2 1 0.055 0.055 0.055 0.055 0.103 0.324 1.588 0.112 2 0.060 0.060 0.060 0.054 0.104 0.322 1.613 0.128 3 0.066 0.064 0.065 0.057 0.101 0.332 1.575 0.097 4 0.081 0.080 0.078 0.053 0.102 0.316 1.551 0.130 5 0.104 0.107 0.107 0.052 0.099 0.302 1.543 0.043 6 0.176 0.170 0.163 0.052 0.105 0.315 1.562 0.122 7 0.294 0.281 0.273 0.053 0.106 0.301 1.609 0.042 8 0.542 0.529 0.494 0.053 0.101 0.318 1.596 0.045 9 0.994 0.941 0.933 0.053 0.100 0.321 1.565 0.039 10 1.831 1.833 1.856 0.054 0.053 0.057 0.053 0.126 11 3.445 3.283 3.294 0.054 0.056 0.053 0.054 0.044 12 out 6.000 4.219 4.134 0.054 0.055 0.059 0.058 0.044 A B C D E F G H Plate 2 1 0.059 0.064 0.064 1.160 0.285 0.100 0.058 0.117 2 0.073 0.069 0.067 1.158 0.274 0.104 0.055 0.098 3 0.073 0.080 0.069 1.141 0.270 0.106 0.062 0.102 4 0.090 0.087 0.079 1.056 0.265 0.107 0.062 0.097 5 0.112 0.115 0.102 1.052 0.263 0.099 0.063 0.098 6 0.171 0.169 0.154 1.013 0.254 0.097 0.063 0.098 7 0.286 0.268 0.261 1.067 0.260 0.097 0.059 0.046 8 0.529 0.496 0.477 1.089 0.263 0.101 0.061 0.043 9 0.943 0.920 0.891 1.051 0.265 0.101 0.063 0.045 10 1.865 1.851 1.758 0.056 0.054 0.060 0.063 0.046 11 3.446 3.320 3.267 0.055 0.058 0.058 0.062 0.044 12 6.000 6.000 6.000 0.056 0.054 0.054 0.058 0.044 NSB 0.055 0.0 0.05 0.060 0.0 0.005 0.10 0.065 1.5 0.010 0.20 0.080 1.9 0.025 0.39 0.106 1.6 0.051 0.78 0.170 3.8 0.115 1.56 0.283 3.7 0.228 3.13 0.522 4.8 0.467 6.25 0.956 3.5 0.901 12.50 1.840 0.8 1.785 25.00 3.341 2.7 3.286 50.00 4.177 1.4 4.122 NSB 0.06233 4.6 0.05 0.07 4.4 0.007 0.10 0.074 7.5 0.012 0.20 0.08533 6.7 0.023 0.39 0.10967 6.2 0.047 0.78 0.16467 5.6 0.102 1.56 0.27167 4.7 0.209 3.13 0.50067 5.3 0.438 6.25 0.918 2.8 0.856 12.50 1.82467 3.2 1.762 25.00 3.34433 2.7 3.282 50.00 6 0.0 5.938 STANDARD CURVE PLATE 1 Concentration (ng/ml) Absorbance 450 nm %CV (A450nm) NSB-corrected absorbance STANDARD CURVE PLATE 2 Concentration (ng/ml) Absorbance 450 nm %CV (A450nm) NSB-corrected absorbance y = 0.1324x1.038 Adjusted standard curves Standard curves 10 10 1 1 R2 = 0.9977 A450-NSB A450-NSB y = 0.1283x1.0296 STANDARD CURVE PLATE 1 0.1 STANDARD CURVE PLATE 2 R2 = 0.9996 STANDARD CURVE PLATE 1 0.1 STANDARD CURVE PLATE 2 Power (STANDARD CURVE PLATE 1) 0.01 0.01 Power (STANDARD CURVE PLATE 2) 0.001 0.01 0.001 0.1 1 10 0.01 100 0.1 1 10 100 ng/ml Vtg ng/ml Vtg SAMPLES PLATE 1 Sample ID Dilution factor Absorbance 450 nm %CV (A450nm) NSB-corrected absorbance Calculated concentration Corrected for dilution factor PJ 10 0.055 2.8 0.000 0.002 0.02 PQ 10 0.052 1.1 -0.003 #NUM! #NUM! PB 10 0.053 0.0 -0.002 #NUM! #NUM! PS 10 0.054 0.0 -0.001 #NUM! #NUM! WJ 10 0.103 1.5 0.048 0.346 3.46 WQ 10 0.102 2.9 0.047 0.341 3.41 WB 10 0.102 3.1 0.047 0.344 3.44 WS 10 0.055 2.8 0.000 #NUM! #NUM! PM 50 0.326 1.6 0.271 2.103 105.17 SAMPLES PLATE 2 Sample ID Dilution factor Absorbance 450 nm %CV (A450nm) NSB-corrected absorbance Calculated concentration Corrected for dilution factor BJ 10 1.153 0.9 1.091 9.06 90.57 BQ 10 1.040 2.3 0.978 8.10 80.95 BB 10 1.069 1.8 1.007 8.34 83.40 BS 10 0.056 1.0 -0.007 #NUM! #NUM! WM 100 0.276 2.8 0.214 1.69 169.34 WM 10000 0.103 3.0 0.041 0.31 3089.53 WM 1000000 0.058 6.0 -0.004 #NUM! #NUM! WA 100 0.261 2.2 0.198 1.57 156.59 WA 10000 0.101 5.2 0.039 0.29 2908.65 PM PM 5000 500000 1.592 0.112 1.2 13.8 1.537 0.057 12.742 0.419 63711.49 ###### WA 1000000 0.063 0.9 0.000 0.00 2178.34 WR 100 0.263 1.0 0.200 1.58 158.22 PA 50 0.311 2.5 0.256 1.983 99.13 PA 5000 1.552 0.6 1.497 12.398 61991 WR 10000 0.100 2.3 0.037 0.28 2805.44 WR 1000000 0.061 3.3 -0.001 #NUM! #NUM! PA 500000 0.098 48.9 0.043 0.314 ###### PR 50 0.313 3.4 0.258 2.001 100.07 *** Page 56 PR 5000 1.590 1.4 1.535 12.725 63625.44 PR 500000 0.042 7.1 -0.013 #NUM! #NUM! Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA E: INDIVIDUAL RAW DATA LAB 1 Day 1 A B C D E F G H Plate 1 1 0.052 0.052 0.054 0.052 0.051 0.059 0.051 0.053 2 0.056 0.057 0.056 0.051 0.052 0.066 0.054 0.054 3 0.063 0.068 0.063 0.051 0.051 0.065 0.056 0.052 4 0.081 0.079 0.078 0.108 0.098 0.095 0.053 0.050 5 0.111 0.107 0.107 0.104 0.096 0.088 0.051 0.051 6 0.176 0.170 0.167 0.105 0.096 0.088 0.051 0.048 7 0.300 0.284 0.287 0.358 0.273 6.000 4.106 0.120 8 0.579 0.553 0.543 0.349 0.276 4.337 3.905 0.119 9 1.097 1.033 0.993 0.349 0.270 4.613 4.023 0.118 10 2.065 2.016 1.930 1.596 1.195 0.052 0.056 0.054 11 3.593 3.429 3.455 1.568 1.230 0.055 0.055 0.052 12 6.000 6.000 4.486 1.618 1.274 0.056 0.052 0.053 A B C D E F G H Plate 2 1 0.052 0.053 0.053 0.059 2.637 0.081 0.050 0.050 2 0.058 0.059 0.056 0.049 2.565 0.081 0.050 0.043 3 0.063 0.065 0.060 0.049 2.503 0.078 0.050 0.042 4 0.078 0.081 0.079 0.103 4.070 1.198 0.063 0.044 5 out 0.125 0.107 0.102 0.102 4.783 1.135 0.062 0.042 6 0.174 0.166 0.163 0.102 4.519 1.152 0.062 0.041 7 0.297 0.276 0.263 0.339 4.560 4.606 1.246 0.043 8 0.556 0.515 0.492 0.334 5.560 4.519 1.195 0.042 9 1.034 0.985 0.953 0.338 5.083 4.481 1.195 0.042 10 2.002 1.911 1.820 1.562 0.054 0.052 0.052 0.044 11 3.555 3.452 3.389 1.620 0.055 0.054 0.053 0.041 12 6.000 4.860 4.258 1.655 0.057 0.053 0.053 0.041 A B C D E F G H Plate 1 1 0.056 0.055 0.052 0.054 0.062 3.096 0.098 0.057 2 0.061 0.058 0.058 0.052 0.055 3.044 0.091 0.057 3 0.067 0.068 0.063 0.052 0.053 2.955 0.088 0.059 4 0.083 0.078 0.078 0.107 0.094 6.000 1.342 0.073 5 0.114 0.109 0.105 0.107 0.088 4.801 1.339 0.074 6 0.175 0.168 0.163 0.109 0.087 4.217 1.322 0.069 7 0.309 0.290 0.295 0.334 0.252 6.000 4.624 1.538 8 0.568 0.559 0.531 0.342 0.252 4.431 6.000 1.538 9 1.043 1.036 1.009 0.344 0.258 4.430 4.462 1.523 10 1.955 1.968 1.894 1.593 1.195 1.552 0.350 0.113 11 3.455 3.514 3.390 1.627 1.162 1.572 0.345 0.115 12 4.574 4.398 4.796 1.714 1.232 1.546 0.363 0.113 A B C D E F G H Plate 2 1 0.053 0.054 0.059 0.051 0.056 0.052 0.051 0.052 2 0.061 0.055 0.061 0.050 0.054 0.050 0.049 0.052 3 0.066 0.064 0.068 out 0.061 0.054 0.052 0.051 0.054 4 0.087 0.079 0.077 0.100 0.087 0.052 0.050 0.050 5 0.115 0.108 0.109 0.114 0.083 0.054 0.050 0.054 6 0.170 0.163 0.158 0.098 0.083 0.054 0.050 0.050 7 0.298 0.279 0.277 0.313 5.264 4.264 0.125 0.054 8 0.519 0.501 0.498 0.313 4.565 4.335 0.123 0.055 9 0.967 0.943 0.927 0.317 4.611 4.286 0.123 0.056 10 1.823 1.829 1.710 1.578 0.380 0.121 0.064 0.057 11 3.635 3.368 3.274 1.642 0.388 0.123 0.072 0.055 12 6.000 6.000 6.000 1.603 0.396 0.125 0.068 0.055 Day 2 Standard curves Day 1 and 2: y = 0.1443x1.0271 Adjusted standard curves y = 0.1413x1.0303 Adjusted standard curves R2 = 0.9986 10 R2 = 0.9984 10 y = 0.1354x1.0401 STANDARD CURVE PLATE 1 0.1 STANDARD CURVE PLATE 2 Power (STANDARD CURVE PLATE 1) 0.01 R2 = 0.9985 1 A450-NSB A450-NSB y = 0.1344x1.0253 R2 = 0.9978 1 STANDARD CURVE PLATE 1 0.1 STANDARD CURVE PLATE 2 Power (STANDARD CURVE PLATE 1) 0.01 Power (STANDARD CURVE PLATE 2) Power (STANDARD CURVE PLATE 2) 0.001 0.01 0.1 1 10 0.001 0.01 100 0.1 1 10 100 ng/ml Vtg ng/ml Vtg *** Page 57 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA LAB 2 Day 1 A B C D E F G H A B C D E F G H Day 2 A B C D E F G H A B C D E F G H Plate 1 1 0.090 0.092 0.093 0.093 0.095 4.648 0.136 0.105 2 0.116 0.145 0.112 0.104 0.093 4.378 0.140 0.108 3 0.127 0.117 0.112 0.089 0.092 4.334 0.134 0.089 4 0.150 0.133 0.136 0.165 0.138 7.531 1.507 0.110 5 0.201 0.177 0.181 0.173 0.152 7.521 1.561 0.116 6 0.272 0.254 0.236 0.150 0.139 7.541 1.548 0.110 7 0.424 0.399 0.385 0.378 0.354 7.601 7.433 1.608 8 0.686 0.647 0.647 0.380 0.341 7.531 7.452 1.566 9 1.290 1.235 1.176 0.373 0.344 7.452 7.511 1.506 10 2.369 2.279 2.206 1.607 1.395 11 4.617 4.473 4.317 1.602 1.443 12 7.531 7.581 7.601 1.615 1.454 Plate 2 1 0.109 0.090 0.126 0.090 0.097 0.306 0.094 2 0.105 0.103 0.100 0.088 0.112 0.317 0.098 3 0.111 0.114 0.111 0.089 0.103 1.252 0.103 4 0.143 0.138 0.145 0.161 0.201 0.092 0.096 5 0.189 0.181 0.182 0.160 0.148 0.091 0.098 6 0.296 0.276 0.264 0.158 0.169 0.091 0.091 7 0.473 0.455 0.430 0.475 7.683 7.395 0.204 8 0.821 0.785 0.743 0.480 7.672 7.462 0.203 9 1.550 1.489 1.430 0.494 7.632 7.414 0.347 10 2.850 2.733 2.652 2.350 11 5.743 5.396 5.614 2.385 12 7.511 7.601 7.591 2.456 Plate 1 1 0.080 0.303 0.075 0.104 0.101 3.910 0.185 0.117 2 0.316 0.195 0.090 0.131 0.223 3.761 0.263 0.176 3 0.130 0.111 0.119 0.093 0.095 3.935 0.129 0.251 4 0.139 0.997 0.811 0.161 0.162 8.583 1.773 0.117 5 0.193 0.180 0.177 0.163 0.148 7.109 1.738 0.112 6 0.276 0.259 0.249 0.152 0.149 6.962 1.700 0.115 7 0.454 0.446 0.442 0.454 0.386 8.481 7.309 2.050 8 0.874 0.802 0.776 0.469 0.373 7.611 6.639 1.926 9 1.597 1.427 1.420 0.472 0.381 9.175 6.495 1.973 10 3.043 2.707 2.693 2.259 1.836 11 5.478 5.111 4.935 2.255 1.898 12 7.906 7.652 7.704 2.422 1.745 Plate 2 1 0.093 0.128 0.091 0.091 0.089 0.105 0.092 2 0.105 0.110 0.125 0.096 0.090 0.101 0.106 3 0.142 0.116 0.112 0.101 0.093 0.093 0.110 4 0.153 0.135 0.145 0.181 0.125 0.092 0.093 5 0.190 0.180 0.176 0.169 0.124 0.102 0.098 6 0.285 0.283 0.221 0.169 0.125 0.095 0.095 7 0.445 0.421 0.400 0.482 7.873 7.501 0.258 8 0.797 0.728 0.699 0.497 7.190 7.423 0.255 9 1.401 1.299 1.247 0.472 7.581 7.414 0.305 10 2.819 2.571 2.357 2.258 11 4.716 4.453 4.424 2.253 12 7.511 7.611 7.601 2.265 Standard curves Day 1 and 2: y = 0.2149x0.915 Adjusted standard curves y = 0.2551x0.9271 Adjusted standard curves R2 = 0.9992 10 R2 = 0.9988 10 1.0206 y = 0.2094x y = 0.2218x0.9439 R2 = 0.9993 STANDARD CURVE PLATE 1 0.1 STANDARD CURVE PLATE 2 A450-NSB A450-NSB R2 = 0.9986 1 1 Power (STANDARD CURVE PLATE 1) 0.01 STANDARD CURVE PLATE 1 0.1 STANDARD CURVE PLATE 2 Power (STANDARD CURVE PLATE 1) 0.01 Power (STANDARD CURVE PLATE 2) Power (STANDARD CURVE PLATE 2) 0.001 0.01 0.1 1 10 0.001 0.01 100 0.1 1 10 100 ng/ml Vtg ng/ml Vtg *** Page 58 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA LAB 3 Day 1 A B C D E F G H Plate 1 1 0.060 0.061 0.061 0.059 0.059 1.591 0.076 0.061 2 0.060 0.060 0.063 0.059 0.059 1.461 0.133 0.061 3 0.063 0.064 0.065 0.060 0.061 1.428 0.077 0.062 4 0.067 0.069 0.071 0.080 0.075 4.000 0.584 0.067 5 0.077 0.077 0.080 0.077 0.073 4.000 0.590 0.066 6 0.098 0.097 0.094 0.076 0.070 4.000 0.577 0.069 7 0.137 0.131 0.132 0.157 0.107 4.000 4.000 0.538 8 0.217 0.209 0.204 0.158 0.145 4.000 4.000 0.535 9 0.370 0.347 0.343 0.161 0.105 4.000 4.000 0.555 10 0.667 0.636 0.631 0.632 0.335 0.061 0.061 0.062 11 1.253 1.180 1.177 0.632 0.342 0.062 0.062 0.060 12 2.271 2.179 2.197 0.694 0.346 0.061 0.062 0.062 A B C D E F G H Plate 2 1 0.064 0.059 0.066 0.064 0.064 0.065 0.078 0.069 2 0.066 0.063 0.067 0.066 0.062 0.065 0.078 0.072 3 0.068 0.064 4 0.076 0.070 0.075 0.089 0.066 0.069 0.080 0.073 5 0.084 0.083 0.086 0.087 0.072 0.067 0.084 0.070 6 0.107 0.104 0.109 0.088 0.069 0.069 0.084 0.072 7 0.151 0.150 0.150 0.179 4.000 2.655 0.097 0.067 8 0.248 0.233 0.259 0.180 4.000 2.536 0.097 0.075 9 0.426 0.418 0.415 0.185 4.000 2.532 0.099 0.070 10 0.773 0.732 0.691 0.717 0.065 0.066 0.067 0.067 11 1.321 1.345 1.388 0.715 0.064 0.071 0.072 0.071 12 2.396 2.277 2.435 0.666 0.074 0.069 0.072 0.074 Day 2 0.063 0.064 0.068 0.084 0.069 A B C D E F G H Plate 1 1 0.058 0.053 0.056 0.060 0.054 1.350 0.072 0.067 2 0.056 0.053 0.056 0.062 0.053 1.191 0.071 0.058 3 0.057 0.057 0.057 0.056 0.059 1.222 0.068 0.056 4 0.062 0.061 0.062 0.074 0.065 4.000 0.605 0.064 5 0.075 0.072 0.073 0.075 0.074 4.000 0.616 0.071 6 0.097 0.093 0.094 0.078 0.065 4.000 0.639 0.067 7 0.140 0.135 0.138 0.161 0.113 4.000 4.000 0.574 8 0.230 0.209 0.211 0.160 0.122 4.000 4.000 0.566 9 0.386 0.345 0.368 0.160 0.109 4.000 4.000 0.562 10 0.759 0.710 0.699 0.654 0.374 0.058 0.054 0.058 11 1.380 1.259 1.253 0.667 0.377 0.056 0.054 0.057 12 2.501 2.383 2.391 0.718 0.425 0.057 0.056 0.056 Plate 2 1 0.069 0.058 0.062 0.060 0.061 0.071 0.073 0.087 2 A B C D E F G H 0.059 0.064 0.061 0.073 0.075 0.077 0.083 3 0.072 0.067 0.067 0.061 0.068 0.075 0.083 0.098 4 0.079 0.074 0.076 0.089 0.068 0.069 0.067 0.083 5 0.094 0.083 0.087 0.090 0.066 0.074 0.071 0.087 6 0.133 0.116 0.124 0.092 0.070 0.073 0.067 0.079 7 0.181 0.189 0.176 0.211 4.000 2.321 0.098 0.087 8 0.294 0.258 0.290 0.191 4.000 2.783 0.109 0.088 9 0.478 0.494 0.482 0.188 4.000 2.615 0.103 0.085 10 0.999 0.950 0.937 0.771 0.069 0.076 0.082 0.082 11 1.734 1.598 1.862 0.864 0.061 0.066 0.074 0.075 2.640 2.797 0.885 0.061 0.070 0.079 0.084 12 Standard curves Day 1 and 2: Adjusted standard curves y = 0.0491x1.0076 Adjusted standard curves y = 0.0461x0.9962 R2 = 0.9989 R2 = 0.9997 10 10 y = 0.0704x0.9712 y = 0.0553x0.9793 R2 = 0.999 STANDARD CURVE PLATE 1 0.1 STANDARD CURVE PLATE 2 Power (STANDARD CURVE PLATE 1) 0.01 R2 = 0.9971 1 A450-NSB A450-NSB 1 STANDARD CURVE PLATE 1 0.1 STANDARD CURVE PLATE 2 Power (STANDARD CURVE PLATE 1) 0.01 Power (STANDARD CURVE PLATE 2) Power (STANDARD CURVE PLATE 2) 0.001 0.01 0.1 1 10 100 0.001 0.01 0.1 1 10 100 ng/ml Vtg ng/ml Vtg *** Page 59 Biosense Laboratories AS Inter-Laboratory validation FHM Vtg ELISA LAB 4 Day 1 A B C D E F G H A B C D E F G H Day 2 A B C D E F G H A B C D E F G H Plate 1 1 0.068 0.049 0.046 0.044 0.087 0.477 0.060 0.164 2 0.548 0.046 0.048 0.057 0.082 0.401 0.051 0.055 3 0.055 0.049 0.345 0.046 0.086 0.649 0.068 0.105 4 0.223 0.052 0.052 0.064 0.060 3.550 0.454 0.108 5 0.059 0.061 0.060 0.067 0.057 3.439 0.447 0.078 6 0.074 0.196 0.088 0.055 0.090 3.551 0.472 0.070 7 0.107 0.147 0.122 0.083 0.112 3.691 4.000 0.374 8 0.166 0.280 0.180 0.126 0.112 3.731 4.000 0.310 9 0.366 0.352 0.361 0.107 0.204 3.659 4.000 0.383 10 0.546 0.516 0.483 0.407 0.357 11 1.173 0.934 1.070 0.370 0.223 12 1.493 2.003 1.745 0.389 0.327 0.056 0.053 0.073 Plate 2 1 0.146 0.137 0.302 0.155 1.143 0.139 0.263 2 0.075 0.150 0.150 0.088 1.807 2.836 2.283 3 0.104 0.183 0.119 0.106 0.266 0.264 2.297 4 0.102 0.132 0.090 0.146 0.203 1.453 0.101 5 0.135 0.165 0.171 0.092 0.269 0.216 1.420 6 0.155 0.361 0.192 0.071 0.225 0.231 0.186 7 0.161 0.384 0.399 0.214 3.543 1.538 1.133 8 0.209 0.349 0.363 0.176 3.702 1.793 0.113 9 0.457 0.404 0.444 0.140 3.576 1.132 0.078 10 0.594 0.674 0.696 0.440 11 1.212 1.082 1.128 0.375 12 1.900 1.736 2.026 0.394 0.203 0.116 0.065 Plate 1 1 0.447 0.381 0.382 0.168 0.206 3.660 0.291 0.214 2 0.393 0.330 0.327 0.152 0.203 3.678 0.244 2.283 3 0.200 0.214 0.220 0.167 0.366 3.683 0.223 0.279 4 0.309 0.313 0.183 0.245 0.240 3.952 2.245 3.900 5 0.240 0.288 0.265 0.364 0.268 3.944 2.247 0.250 6 0.472 0.524 0.604 0.307 1.087 3.968 2.194 0.245 7 0.569 0.705 0.637 0.746 0.414 4.000 4.000 2.805 8 1.628 0.940 1.373 0.614 0.483 4.000 4.000 2.745 9 1.935 1.620 1.622 0.596 1.825 4.000 4.000 2.679 10 2.677 2.626 2.510 2.570 1.434 11 3.800 3.740 3.709 2.380 1.514 12 4.000 4.000 4.000 2.216 1.394 0.157 0.198 0.368 Plate 2 1 2.064 0.235 0.927 0.087 0.095 0.074 0.155 2 0.287 0.080 0.122 0.190 0.133 0.097 0.071 3 0.139 0.913 3.789 0.072 0.137 0.069 0.065 4 0.173 0.224 1.950 0.160 0.105 0.065 0.139 5 0.242 0.230 0.215 0.158 0.090 0.092 0.104 6 0.361 0.536 0.791 0.157 0.088 0.074 0.059 7 0.440 0.424 0.408 3.699 4.000 4.000 0.230 8 0.763 0.721 0.674 2.045 4.000 4.000 0.224 9 1.108 1.221 1.189 0.451 4.000 4.000 1.087 10 2.101 2.238 2.213 1.973 11 3.582 3.506 3.445 1.790 12 4.000 4.000 4.000 1.790 0.091 0.231 0.144 Standard curves Day 1 and 2: Adjusted standard curves 2 R = 0.9936 10 y = 0.2757x0.8992 Adjusted standard curves y = 0.0376x1.0542 R2 = 0.9814 10 0.8386 y = 0.2315x y = 0.0646x0.8492 R2 = 0.9935 0.1 0.01 R2 = 0.9978 1 A450-NSB A450-NSB 1 STANDARD CURVE PLATE 1 0.1 STANDARD CURVE PLATE 2 Power (STANDARD CURVE PLATE 1) 0.01 STANDARD CURVE PLATE 1 Power (STANDARD CURVE PLATE 2) STANDARD CURVE PLATE 2 0.001 0.01 0.1 1 ng/ml Vtg 10 Power (STANDARD CURVE PLATE 1) 100 0.001 0.01 Power (STANDARD CURVE PLATE 2) 0.1 1 10 100 ng/ml Vtg *** Page 60