Key Terms:
advertisement

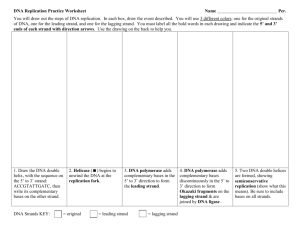
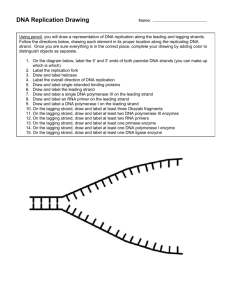
Bio102: Introduction to Cell Biology and Genetics Replication and Chromosomes Key Terms: Chromatin Histones Chromosome Homologous pair N, 2N Origin Initiator Protein Helicase Topoisomerase Primase Leading Strand Lagging Strand Okazaki Fragment Ligase DNA Polymerase III DNA Polymerase I Replication Bubble Replication Fork Key Figures: Figures 16.9, 16.12, 16.13, 16.14, 16.15, 16.16, 16.17, 16.21 Key Questions: What features of Watson and Crick's DNA structure suggested a model for how it would replicate? What is a homologous pair of chromosomes? List the enzymes and other proteins involved in DNA replication and briefly describe their functions. Draw a replication bubble. Label 5' and 3' ends. For each of the two replication forks, show the leading strand, lagging strand, helicase, RNA primers, Okazaki fragments and two DNA polymerase III molecules Lecture Outline: In a cell, DNA is highly condensed. Associated with many proteins as chromatin two alleles of each gene, so two homologous pairs of chromosomes (may have different alleles) Each species has a constant number of unique chromosomes (N). Diploid cells are 2N. in humans, N=23. So diploid human cells have 46 chromosomes or 23 homologous pairs DNA Polymerase III is the major enzyme to connect nucleotides Origin of Replication is bound by Initiator Protein recruits Helicase to ‘unzip’ helix creates a replication bubble with two equivalent replication forks causes tension; released by topoisomerases DNA Polymerases can’t start polymerizing. Need a 3’ end to add on to Primase makes short RNA primers that can be extended by DNA Polymerase III DNA Polymeraes can only add to a 3’ end, not a 5’ end polymerization of the two strands at one fork move in different overall directions leading (continuous) strand vs. lagging (noncontinuous) strand multiple priming events needed on the lagging strand making Okazaki fragments DNA Polymerase I removes residual RNA and fills in with DNA Ligase connects adjacent Okazaki fragments with a sugar-phosphate connection