Supporting Information to Wylie, D
advertisement

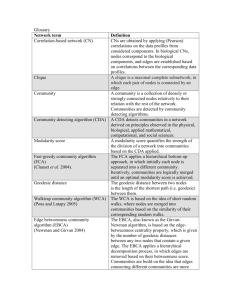
Supporting Information to Wylie, D.C. and W.M. Getz. Sick and Edgy: Walk-Counting as a Metric of Epidemic Spreading on Networks. Section 1. Structure Metrics I Metric Degree Distribution [2] Assortativity [38-40] Transitivity / Clustering Coefficient [1, 4] Characteristic Path Length / Radius / Diameter [4, 41] Scope Local TwoNode Threenode Global Random Graph Relatively Homogenous Not assortative Small-World Very Homogenous Not assortative Scale-Free Power-law (very skewed) Not assortative Not clustered Highly clustered Not clustered O(log(n)) Dependent on Rewiring probability p: O(n) at p=0; becomes O(log(n)) at small p>0 None O(log(n)) (very short) Community Structure Global Not much* None [42-47] Table SI.1. Characteristics of canonical network structures as measured by the most commonly used structural metrics. Many different metrics have been suggested for consideration of the “structure” of a graph/network – e.g., degree distribution, transitivity/clustering coefficients, presence of certain small subgraphs (“motifs”), diameter, etc. [1-3]. Some of the most commonly considered such metrics merit further discussion here, and will be described in order of increasing “globality.” Perhaps the most familiar, and most local, measure of network structure is the degree distribution [2]. This measure may be described as the probability distribution for the quantity k representing the number of links to other nodes of a node picked from the network in question at random, with uniform probability. Of particular interest in this regard are scale-free networks [5], which are defined by a power-law degree distribution, P(k)~k-γ, with, in many relevant cases, the exponent γ<3 [2, 3]. This sort of distribution may thus be so skewed that all moments above the first diverge, with the consequence that the minimum transmissibility for possibility of epidemic outbreak declines to zero [21, 22, 27]. There is a greater variety of slightly less local network metrics which might collectively be described as “few-node correlation” measures. For example, the assortativity (or dissortativity) of a network considers the properties of nodes taken in pairs, measuring the correlation between the degree of a node and that of its neighbors [38-40]. The transitivity [3] (or, alternately, the clustering coefficient [3, 4]) considers triples of nodes by monitoring the prevalence of triangles (i.e., collections of three nodes which are completely connected) in the network. The small-world networks of Watts and Strogatz [4] have finite (rewiring probability p-dependent) transitivities/clustering coefficients at all sizes [2]. Milo et. al. [48] proposed the examination of networks for the repeated presence of certain few-node substructures (motifs) in significant excess of what might be expected from a suitable randomized null-model. Any of the three network types taken here as canonical might serve as such an appropriate null-model, depending on the specific network to be analyzed for motifs. In contrast to the above, the characteristic path length [4] (and similar measures such as the diameter and radius [41]) of a network, defined as the longest distance between any two nodes, is a truly global metric and cannot generally be computed by considering only small components of the network one-by-one. Finally, some networks may be divided into a set of subnetworks, or communities, such that within each community, the density of interconnections between nodes is appreciably higher than the connection density among nodes taken from distinct communities [42]. The problem of subdividing networks into communities is clearly global in nature; a number of methods for approaching this task have been suggested [42-47]. Section 2. Structure Metrics II – Matrix Methods Various matrices may be associated with networks, and there have been many results indicating that the linear algebraic properties of these matrices can contain useful structural information [37, 49-54]. For the purposes of this paper, one of the most important such results is also one of the simplest, namely that the number of k-walks (i.e., walks of k steps) from node i to node j in a given network G is given by (Ak)ij [49], where the adjacency matrix A = A(G) is defined by Aij = 1 if there is an edge connecting node i to node j in G, and Aij = 0 otherwise. This may be proven so by a simple induction; after noting that the base case for k = 1 is trivially true, consider: 1) A k 1 ij A k iq Aqj q A k q| q j iq where, in the second line, a indicates the set of integers q such that node q neighbors node j (symbolically represented by qj). Thus Equation (16) indicates that the number of (k+1)-walks from i to j is equal to the sum of the numbers of k-walks from i to q over all neighbors q of j, completing the inductive argument, since each (k+1)-walk from i to j consists of a k-walk from i to a neighbor q of j, followed by the step from q to j. Much more attention has been given to spectral properties of both the adjacency matrix as defined above and the Laplacian matrix, for which alternative definitions exist [51]. The simplest version, the combinatorial Laplacian, may be defined by the component equation Lij = Diij - Aij, where Di is the degree of node i. The spectra of both the adjacency matrix and the Laplacian matrix have particularly nice properties for regular networks (that is, networks in which all nodes have the same degree) [50]. An oft-mentioned property of the Laplacian matrix is that the multiplicity of the smallest eigenvalue (0) is equal to the number of connected components of the network. For a connected network, it is known that the magnitude of the second-smallest eigenvalue of L is related to the number of edges that must be removed to split the network into pieces [50]. With regard to the “canonical” network structures, a great deal has been learned regarding their adjacency matrix spectrum [37, 52, 54, 55]. The most relevant features from the point of view of this paper pertain to the largest (in magnitude) eigenvalues. Scale-free networks generally have larger principal eigenvalues (scaling with the fourth root of the number of nodes) than do the other two canonical network species, which, for fixed average node degree, both feature principal eigenvalues asymptotically independent of network size [37]. However, Watts-Strogatz small-world networks generally have the largest number of eigenvalues clustered at approximately the same value as the principal eigenvalue, while Barabasi-Albert scale-free networks generally have the largest separation between the magnitude of the principal eigenvalue and the remainder of the spectrum [37]. This last point is related to the high degree of clustering present in small-world networks. Networks with community structure also tend to have multiple eigenvalues clustered just below the principal eigenvalue, and in fact this property is exploited by many of the algorithms designed to detect communities in networks [3, 44, 46]. Section 3. Construction of Adjustable Network Topologies Network construction is divided into five phases: 1.) During the first phase, C distinct BA scale-free networks of (n/C) nodes with average degree dsf (<d) are assembled. 2.) During the second phase, each remaining pair of edges within each network is connected with probability ((d-dsf)/((n/C)-dsf-1)), so that the average degree of the nodes is d in each of the C networks. 3.) The third phase of construction involves rearranging edges within each of the C networks so as to adjust assortativity without altering the degree distribution; this is done by an algorithm similar to that described in Xulvi-Brunet and Sokolov [40]. Described briefly, this method involves repeatedly selecting, a particular number of times denoted by Nsort-swap, pairs of node-disjoint edges (both within the same network) at random and breaking them. The four nodes involved will then be rewired with two new edges, chosen either at random (with probability 1-|psort|) or (with probability |psort|) according to a scheme to adjust assortativity. If the latter option is chosen, the two new edges will be either (if psort>0) a new edge connecting the two highest degree nodes of the four involved in the original edges, as well as a connection between the two remaining lower degree nodes, or (if psort<0) a new edge connecting the two extremal degree nodes together, along with a new connection between the two nodes of middling degree. If either of the putative new edges would be identical to a preexisting edge, all changes are aborted and the iteration begun anew (unless a certain threshold (25*Nsort-swap) number of total attempts have been made, successfully or unsucessfully, in which case no further iterations are pursued). 4.) The C networks (which will now be referred to as communities) are finally connected to each other during the fourth phase. This phase consists of an iterative procedure in which, at each iteration, one edge is chosen from each of two distinct communities. These two connections are broken and replaced by two new connections bridging the two communities in question. One of the new connections connects the first node of the broken edge in one community to either the first or second node (50% chance of either) of the broken edge in the other community, while the other new connection connects the remaining two nodes that were involved in the broken edges to each other. (If either of the resulting new connections would be the same as a preexisting connection, the procedure is begun anew.) Note that this procedure leaves the degree of all nodes invariant, and hence does not affect the degree distribution (though it will generally change the assortativity of the network). The selected number of iterations executed, denoted by Ncluster-swap, then controls both how strongly clustered the resulting network is and how well-defined the various communities will be. 5.) The final phase of “adjustable network” construction consists of a series of edge swaps between edges which connect distinct communities in order to further adjust the assortativity of the network, again in a manner very similar to that of Xulvi-Brunet [40]. If the network is composed of C>1 communities then the following procedure is then repeated iteratively a number of times, denoted by Ncluster-sort-swap: randomly choose two edges which bridge distinct communities. With probability |psort|, break these two edges and replace them with one edge connecting the two highest degree nodes together and one edge connecting the remaining (lower degree) nodes (if psort>0), or with one edge connecting the two nodes of middling degree together and one connecting the nodes of extremal degree together (if psort<0). Reject the rearrangement and begin the iteration anew (unless 25*Ncluster-sort-swap total iteration attempts have been made, in which case no further iterations are pursued) if either of the new edges would not bridge distinct communities or if either of the new connections would be the same as a preexisting connection.