We illustrate the variable selection methods on some data on the 50
advertisement

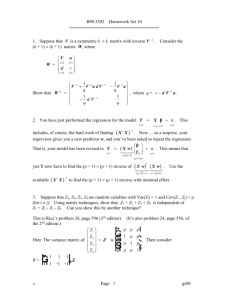
We illustrate the variable selection methods on some data on the 50 states - the variables are population estimate as of July 1, 1975; per capita income (1974); illiteracy (1970, percent of population); life expectancy in years (1969-71); murder and non-negligent manslaughter rate per 100,000 population (1976); percent high-school graduates (1970); mean number of days with min temperature < 32 degrees (1931- 1960) in capital or large city; and land area in square miles. The data was collected from US Bureau of the Census. We will take life expectancy as the response and the remaining variables as predictors - a fix is necessary to remove spaces in some of the variable names. > data(state) > help(state) > statedata<data.frame(state.x77,row.names=state.abb,check.names=T) > g <- lm(Life.Exp~., data=statedata) > summary(g) ##### outputs ###### Coefficients: Estimate (Intercept) 7.09e+01 Population 5.18e-05 Income -2.18e-05 Illiteracy 3.38e-02 Murder -3.01e-01 HS.Grad 4.89e-02 Frost -5.74e-03 Area -7.38e-08 Std. Error 1.75e+00 2.92e-05 2.44e-04 3.66e-01 4.66e-02 2.33e-02 3.14e-03 1.67e-06 t value 40.59 1.77 -0.09 0.09 -6.46 2.10 -1.82 -0.04 Pr(>|t|) < 2e-16 0.083 0.929 0.927 8.7e-08 0.042 0.075 0.965 Residual standard error: 0.745 on 42 degrees of freedom Multiple R-Squared: 0.736, Adjusted R-squared: 0.692 F-statistic: 16.7 on 7 and 42 degrees of freedom,p-value:2.53e-10 Which predictors should be included - can you tell from the p-values? Looking at the coefficients, can you see what operation would be helpful? Does the murder rate decrease life expectancythat’s obvious a priori, but how should these results be interpreted? We illustrate the backward method - at each stage we remove the predictor with the largest p-value over 0.05: > g <- update(g, .~. - Area) > summary(g) Coefficients: Estimate Std. Error (Intercept) 7.10e+01 1.39e+00 Population 5.19e-05 2.88e-05 Income -2.44e-05 2.34e-04 Illiteracy 2.85e-02 3.42e-01 Murder -3.02e-01 4.33e-02 HS.Grad 4.85e-02 2.07e-02 Frost -5.78e-03 2.97e-03 > g <- update(g, .~. - Illiteracy) > summary(g) t value 51.17 1.80 -0.10 0.08 -6.96 2.35 -1.94 Pr(>|t|) < 2e-16 0.079 0.917 0.934 1.5e-08 0.024 0.058 Coefficients: (Intercept) Population Income Murder HS.Grad Frost Estimate 7.11e+01 5.11e-05 -2.48e-05 -3.00e-01 4.78e-02 -5.91e-03 Std. Error 1.03e+00 2.71e-05 2.32e-04 3.70e-02 1.86e-02 2.47e-03 t value 69.07 1.89 -0.11 -8.10 2.57 -2.39 Pr(>|t|) < 2e-16 0.066 0.915 2.9e-10 0.014 0.021 > g <- update(g, .~. - Income) > summary(g) Coefficients: Estimate (Intercept) 7.10e+01 Population 5.01e-05 Murder -3.00e-01 HS.Grad 4.66e-02 Frost -5.94e-03 Std.Error 9.53e-01 2.51e-05 3.66e-02 1.48e-02 2.42e-03 t value 74.54 2.00 -8.20 3.14 -2.46 Pr(>|t|) < 2e-16 0.0520 1.8e-10 0.0030 0.0180 > g <- update(g, .~. - Population) > summary(g) Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 71.03638 0.98326 72.25 <2e-16 Murder -0.28307 0.03673 -7.71 8e-10 HS.Grad 0.04995 0.01520 3.29 0.0020 Frost -0.00691 0.00245 -2.82 0.0070 Residual standard error: 0.743 on 46 degrees of freedom Multiple R-Squared: 0.713, Adjusted R-squared: 0.694 F-statistic: 38 on 3 and 46 degrees of freedom, p-value: 1.63e-12 The final removal of the Population variable is a close call. We may want to consider including this variable if interpretation is aided. Notice that the R2 for the full model of 0.736 is reduced only slightly to 0.713 in the final model. Thus the removal of four predictors causes only a minor reduction in fit. Best subset: Cp Note: Here you might need two packages: leaps and faraway. How to install packages? It is usual to plot Cp against p. We desire models with small p and Cp around or less than p. Now we try the Cp and adjR2 methods for the selection of variables in the State dataset. The default for the leaps() function is the Mallow’s Cp criterion: > library(leaps) > library(faraway) > x <- model.matrix(g)[,-1] > y <- statedata$Life > g <- leaps(x,y) > Cpplot(g) If we get the following Cpplot, what should we do? The models are denoted by indices for the predictors. The competition is between the “456” model i.e. the Frost, HS graduation and Murder model and the model also including Population. Both models are on or below the Cp =p line, indicating good fits. The choice is between the smaller model and the larger model which fits a little better. Some even larger models fit in the sense that they are on or below theCp=p line but we would not opt for these in the presence of smaller models that fit. Smaller models with 1 or 2 predictors are not shown on this plot because their Cp plots are so large. _ > adjr <- leaps(x,y,method="adjr2") > maxadjr(adjr,8) 1456 12456 13456 14567 123456 134567 124567 456 0.713 0.706 0.706 0.706 0.699 0.699 0.699 0.694 We see that the Population, Frost, HS graduation and Murder model has the largest adj R2. The best three-predictor model is in eighth place but the intervening models are not attractive since they use more predictors than the best model. ANOVA in R Consider a model with only one variable, income: >g1 <- lm(Life.Exp~Income, data=statedata) >summary(g1) ## Take a look >anova(g1) ## Analysis of Variance Call: lm(formula = Life.Exp ~ Income, data = statedata) Residuals: Min 1Q Median -2.96547 -0.76381 -0.03428 3Q 0.92876 Max 2.32951 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 6.758e+01 1.328e+00 50.906 <2e-16 *** Income 7.433e-04 2.965e-04 2.507 0.0156 * --Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 1.275 on 48 degrees of freedom Multiple R-Squared: 0.1158, Adjusted R-squared: 0.09735 F-statistic: 6.285 on 1 and 48 DF, p-value: 0.01562 Analysis of Variance Table Response: Life.Exp Df Sum Sq Mean Sq F value Pr(>F) Income 1 10.223 10.223 6.2847 0.01562 * Residuals 48 78.076 1.627 -3 -1.5 -1.0 -2 -0.5 0.0 residuals 0 -1 residuals 0.5 1 1.0 2 1.5 Residual plots in R g <- lm(Life.Exp~., data=statedata) g1<-lm(Life.Exp~Income, data=statedata) par(mfrow=c(1,2)) ## draw two figures plot(g1$fit,g1$res,xlab="Fitted",ylab="residuals") plot(g$fit,g$res,xlab="Fitted",ylab="residuals") ### What is the difference? 70.0 70.5 71.0 Fitted 71.5 72.0 69 70 71 72 Fitted Plot against ŷ . This is the most important diagnostic plot that you can make. If all is well, you should see constant variance in the vertical ( ˆ) direction and the scatter should be symmetric vertically about 0. Things to look for are heteroscedascity (non-constant variance) and nonlinearity (which indicates some change in the model is necessary). Look at the following figures: Residuals vs Fitted plots - the first suggests no change to the current model while the second shows non-constant variance and the third indicates some nonlinearity which should prompt some change in the structural form of the model. You should also plot ˆagainst xi (for predictors that are both in and out of the model). Look for the same things except in the case of plots against predictors not in the model, look for any relationship which might indicate that this predictor should be included. 2 1 0 -3 -2 -1 residuals 0 -1 -2 -3 residuals 1 2 For example: plot(statedata$Popula,g1$res,xlab="Population",ylab="residuals") plot(statedata$Murder,g1$res,xlab="Murder",ylab="residuals") 0 5000 10000 Population 15000 20000 2 4 6 8 Murder 10 12 14