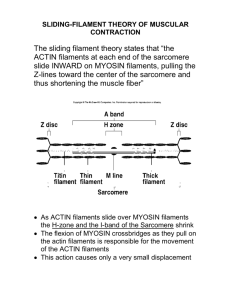
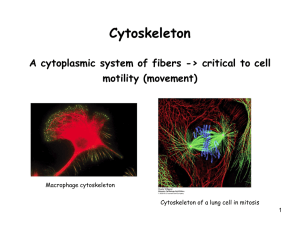
Document
advertisement

11. The Cytoskeleton and Cell Movement The membrane-enclosed organelles discussed in the preceding chapters constitute one level of the organizational substructure of eukaryotic cells. A further level of organization is provided by the cytoskeleton, which consists of a network of protein filaments extending throughout the cytoplasm of all eukaryotic cells. The cytoskeleton provides a structural framework for the cell, serving as a scaffold that determines cell shape and the general organization of the cytoplasm. In addition to playing this structural role, the cytoskeleton is responsible for cell movements. These include not only the movements of entire cells, but also the internal transport of organelles and other structures (such as mitotic chromosomes) through the cytoplasm. Importantly, the cytoskeleton is much less rigid and permanent than its name implies. Rather, it is a dynamic structure that is continually reorganized as cells move and change shape, for example, during cell division. The cytoskeleton is composed of three principal types of protein filaments: actin filaments, intermediate filaments, and microtubules, which are held together and linked to subcellular organelles and the plasma membrane by a variety of accessory proteins. This chapter discusses the structure and organization of each of these three major components of the cytoskeleton, as well as their roles in cell motility, organelle transport, cell division, and other types of cell movements. Structure and Organization of Actin Filaments The major cytoskeletal protein of most cells is actin, which polymerizes to form actin filaments thin, flexible fibers approximately 7 nm in diameter and up to several micrometers in length. Within the cell, actin filaments (also called microfilaments) are organized into higher-order structures, forming bundles or three-dimensional networks with the properties of semisolid gels. The assembly and disassembly of actin filaments, their crosslinking into bundles and networks, and their association with other cell structures (such as the plasma membrane) are regulated by a variety of actin-binding proteins, which are critical components of the actin cytoskeleton. Actin filaments are particularly abundant beneath the plasma membrane, where they form a network that provides mechanical support, determines cell shape, and allows movement of the cell surface, thereby enabling cells to migrate, engulf particles, and divide. Assembly and Disassembly of Actin Filaments Actin was first isolated from muscle cells, in which it constitutes approximately 20% of total cell protein, in 1942. Although actin was initially thought to be uniquely involved in muscle contraction, it is now known to be an extremely abundant protein (typically 5 to 10% of total protein) in all types of eukaryotic cells. Yeasts have only a single actin gene, but higher eukaryotes have several distinct types of actin, which are encoded by different members of the actin gene family. Mammals, for example, have at least six distinct actin genes: Four are expressed in different types of muscle and two are expressed in nonmuscle cells. All of the actins, however, are very similar in amino acid sequence and have been highly conserved throughout the evolution of eukaryotes. Yeast actin, for example, is 90% identical in amino acid sequence to the actins of mammalian cells. The three-dimensional structures of both individual actin molecules and actin filaments were determined in 1990 by Kenneth Holmes, Wolfgang Kabsch, and their colleagues. Individual actin molecules are globular proteins of 375 amino acids (43 kd). Each actin monomer (globular [G] actin) has tight binding sites that mediate head-to-tail interactions with two other actin monomers, so actin monomers polymerize to form filaments (filamentous [F] actin) (Figure 11.2). Figure 11.2. Assembly and structure of actin filaments (A) Actin monomers (G actin) polymerize to form actin filaments (F actin). The first step is the formation of dimers and trimers, which then grow by the addition of monomers to both ends. (B) Structure of an actin monomer. Each monomer is rotated by 166o in the filaments, which therefore have the appearance of a double-stranded helix. Because all the actin monomers are oriented in the same direction, actin filaments have a distinct polarity and their ends (called the plus and minus ends) are distinguishable from one another. This polarity of actin filaments is important both in their assembly and in establishing a unique direction of myosin movement relative to actin, as discussed later in the chapter. The assembly of actin filaments can be studied in vitro by regulation of the ionic strength of actin solutions. In solutions of low ionic strength, actin filaments depolymerize to monomers. Actin then polymerizes spontaneously if the ionic strength is increased to physiological levels. The first step in actin polymerization (called nucleation) is the formation of a small aggregate consisting of three actin monomers. Actin filaments are then able to grow by the reversible addition of monomers to both ends, but one end (the plus end) elongates five to ten times faster than the minus end. The actin monomers also bind ATP, which is hydrolyzed to ADP following filament assembly. Although ATP is not required for polymerization, actin monomers to which ATP is bound polymerize more readily than those to which ADP is bound. As discussed below, ATP binding and hydrolysis play a key role in regulating the assembly and dynamic behavior of actin filaments. Because actin polymerization is reversible, filaments can depolymerize by the dissociation of actin subunits, allowing actin filaments to be broken down when necessary (Figure 11.3). Figure 11.3. Reversible polymerization of actin monomers Actin polymerization is a reversible process, in which monomers both associate with and dissociate from the ends of actin filaments. The rate of subunit dissociation (koff) is independent of monomer concentration, while the rate of subunit association is proportional to the concentration of free monomers and given by C × kon (C = concentration of free monomers). An apparent equilibrium is reached at the critical concentration of monomers (Cc), where koff = Cc × kon. Thus, an apparent equilibrium exists between actin monomers and filaments, which is dependent on the concentration of free monomers. The rate at which actin monomers are incorporated into filaments is proportional to their concentration, so there is a critical concentration of actin monomers at which the rate of their polymerization into filaments equals the rate of dissociation. At this critical concentration, monomers and filaments are in apparent equilibrium. As noted earlier, the two ends of an actin filament grow at different rates, with monomers being added to the fastgrowing end (the plus end) five to ten times faster than to the slow-growing (minus) end. Because ATP-actin dissociates less readily than ADP-actin, this results in a difference in the critical concentration of monomers needed for polymerization at the two ends. This difference can result in the phenomenon known as treadmilling, which illustrates the dynamic behavior of actin filaments (Figure 11.4). Figure 11.4. Treadmilling The minus ends grow less rapidly than the plus ends of actin filaments. This difference in growth rate is reflected in a difference in the critical concentration for addition of monomers to the two ends of the filament. Actin bound to ATP associates with the rapidly growing plus ends, and the ATP bound to actin is then hydrolyzed to ADP. Because ADP-actin dissociates from filaments more readily than ATP-actin, the critical concentration of actin monomers is higher for addition to the minus end than to the plus end of actin filaments. Treadmilling takes place at monomer concentrations intermediate between the critical concentrations for the plus and minus ends. Under these conditions, there is a net dissociation of monomers (bound to ADP) from the minus end, balanced by the addition of monomers (bound to ATP) to the plus end. For the system to be at an overall steady state, the concentration of free actin monomers must be intermediate between the critical concentrations required for polymerization at the plus and minus ends of the actin filaments. Under these conditions, there is a net loss of monomers from the minus end, which is balanced by a net addition to the plus end. Treadmilling requires ATP, with ATP-actin polymerizing at the plus end of filaments while ADP-actin dissociates from the minus end. Although the role of treadmilling in the cell is unclear, it may reflect the dynamic assembly and disassembly of actin filaments required for cells to move and change shape. It is noteworthy that several drugs useful in cell biology act by binding to actin and affecting its polymerization. For example, the cytochalasins bind to the plus ends of actin filaments and block their elongation. This results in changes in cell shape as well as inhibition of some types of cell movements (e.g., cell division following mitosis), indicating that actin polymerization is required for these processes. Another drug, phalloidin, binds tightly to actin filaments and prevents their dissociation into individual actin molecules. Phalloidin labeled with a fluorescent dye is frequently used to visualize actin filaments by fluorescence microscopy. Within the cell, both the assembly and disassembly of actin filaments are regulated by actin-binding proteins (Figure 11.5). The turnover of actin filaments is about 100 times faster within the cell than it is in vitro, and this rapid turnover of actin plays a critical role in a variety of cell movements. The key protein responsible for actin filament disassembly within the cell is cofilin, which binds to actin filaments and enhances the rate of dissociation of actin monomers from the minus end. In addition, cofilin can sever actin filaments, generating more ends and further enhancing filament disassembly. Figure 11.5. Effects of actin-binding proteins on filament turnover Cofilin binds to actin filaments and increases the rate of dissociation of actin monomers (bound to ADP) from the minus end. Cofilin remains bound to the ADP-actin monomers, preventing their reassembly into filaments. However, profilin can stimulate the exchange of bound ADP for ATP, resulting in the formation of ATPactin monomers that can be repolymerized into filaments, including new filaments nucleated by the Arp2/3 proteins. Cofilin preferentially binds to ADP-actin, so it remains bound to actin monomers following filament disassembly and sequesters them in the ADP-bound form, preventing their reincorporation into filaments. However, another actin-binding protein, profilin, can reverse this effect of cofilin and stimulate the incorporation of actin monomers into filaments. Profilin acts by stimulating the exchange of bound ADP for ATP, resulting in the formation of ATPactin monomers, which dissociate from cofilin and are then available for assembly into filaments. Other proteins (Arp2/3 proteins) can serve as nucleation sites to initiate the assembly of new filaments, so cofilin, profilin, and the Arp2/3 proteins (as well as other actin-binding proteins) can act together to promote the rapid turnover of actin filaments and remodeling of the actin cytoskeleton which is required for a variety of cell movements and changes in cell shape. As might be expected, the activities of cofilin, profilin, and Arp2/3 proteins are controlled by a variety of cell signaling mechanisms, allowing actin polymerization to be appropriately regulated in response to environmental stimuli. Organization of Actin Filaments Individual actin filaments are assembled into two general types of structures, called actin bundles and actin networks, which play different roles in the cell (Figure 11.6). Figure 11.6. Actin bundles and networks (B) Schematic organization of bundles and networks. Actin filaments in bundles are crosslinked into parallel arrays by small proteins that align the filaments closely with one another. In contrast, networks are formed by large flexible proteins that crosslink orthogonal filaments. In bundles, the actin filaments are crosslinked into closely packed parallel arrays. In networks, the actin filaments are loosely crosslinked in orthogonal arrays that form threedimensional meshworks with the properties of semisolid gels. The formation of these structures is governed by a variety of actin-binding proteins that crosslink actin filaments in distinct patterns. All of the actin-binding proteins involved in crosslinking contain at least two domains that bind actin, allowing them to bind and crosslink two different actin filaments. The nature of the association between these filaments is then determined by the size and shape of the crosslinking proteins (see Figure 11.6). The proteins that crosslink actin filaments into bundles (called actin-bundling proteins) usually are small rigid proteins that force the filaments to align closely with one another. In contrast, the proteins that organize actin filaments into networks tend to be large flexible proteins that can crosslink perpendicular filaments. These actin-crosslinking proteins appear to be modular proteins consisting of related structural units. In particular, the actin-binding domains of many of these proteins are similar in structure. They are separated by spacer sequences that vary in length and flexibility, and it is these differences in the spacer sequences that are responsible for the distinct crosslinking properties of different actin-binding proteins. There are two structurally and functionally distinct types of actin bundles, involving different actin-bundling proteins (Figure 11.7). Figure 11.7. Actin-bundling proteins Actin filaments are associated into two types of bundles by different actin-bundling proteins. Fimbrin has two adjacent actin-binding domains (ABD) and crosslinks actin filaments into closely packed parallel bundles in which the filaments are approximately 14 nm apart. In contrast, the two separated actin-binding domains of -actinin dimers crosslink filaments into more loosely spaced contractile bundles in which the filaments are separated by 40 nm. Both fimbrin and -actinin contain two related Ca2+binding domains, and -actinin contains four repeated -helical spacer domains. The first type of bundle, containing closely spaced actin filaments aligned in parallel, supports projections of the plasma membrane, such as microvilli (see 11.16). In these bundles, all the filaments have the same polarity, with their plus ends adjacent to the plasma membrane. An example of a bundling protein involved in the formation of these structures is fimbrin, which was first isolated from intestinal microvilli and later found in surface projections of a wide variety of cell types. Fimbrin is a 68-kd protein, containing two adjacent actin-binding domains. It binds to actin filaments as a monomer, holding two parallel filaments close together. The second type of actin bundle is composed of filaments that are more loosely spaced and are capable of contraction, such as the actin bundles of the contractile ring that divides cells in two following mitosis. The looser structure of these bundles (which are called contractile bundles) reflects the properties of the crosslinking protein -actinin. In contrast to fimbrin, -actinin binds to actin as a dimer, each subunit of which is a 102-kd protein containing a single actin-binding site. Filaments crosslinked by -actinin are consequently separated by a greater distance than those crosslinked by fimbrin (40 nm apart instead of 14 nm). The increased spacing between filaments allows the motor protein myosin to interact with the actin filaments in these bundles, which (as discussed later) enables them to contract. The actin filaments in networks are held together by large actin-binding proteins, such as filamin (Figure 11.8). Filamin (also called actin-binding protein or ABP-280) binds actin as a dimer of two 280-kd subunits. The actin-binding domains and dimerization domains are at opposite ends of each subunit, so the filamin dimer is a flexible V-shaped molecule with actin-binding domains at the ends of each arm. As a result, filamin forms cross-links between orthogonal actin filaments, creating a loose three-dimensional meshwork. As discussed in the next section, such networks of actin filaments underlie the plasma membrane and support the surface of the cell. Figure 11.8. Actin networks and filamin Filamin is a dimer of two large (280-kd) subunits, forming flexible V-shaped molecule that crosslinks actin filaments into orthogonal networks. The carboxyterminal dimerization domain is separated from the amino-terminal actin-binding domain by repeated sheet spacer domains. a - Association of Actin Filaments with the Plasma Membrane Actin filaments are highly concentrated at the periphery of the cell, where they form a three-dimensional network beneath the plasma membrane (see Figure 11.6). This network of actin filaments and associated actin-binding proteins (called the cell cortex) determines cell shape and is involved in a variety of cell surface activities, including movement. The association of the actin cytoskeleton with the plasma membrane is thus central to cell structure and function. Red blood cells (erythrocytes) have proven particularly useful for studies of both the plasma membrane (discussed in the next chapter) and the cortical cytoskeleton. The principal advantage of red blood cells for these studies is that they contain no nucleus or internal organelles, so their plasma membrane and associated proteins can be easily isolated without contamination by the various internal membranes that are abundant in other cell types. In addition, human erythrocytes lack other cytoskeletal components (microtubules and intermediate filaments), so the cortical cytoskeleton is the principal determinant of their distinctive shape as biconcave discs. The major protein that provides the structural basis for the cortical cytoskeleton in erythrocytes is the actin-binding protein spectrin, which is related to filamin (Figure 11.10). Figure 11.10. Structure of spectrin Spectrin is a tetramer consisting of two and two chains. Each chain has a single actin-binding domain (ABD) at its amino terminus. Both and chains contain multiple repeats of -helical spacer domains, which separate the two actin-binding domains of the tetramer. The chain has two Ca2+ binding domains at its carboxy terminus. Erythrocyte spectrin is a tetramer consisting of two distinct polypeptide chains, called and , with molecular weights of 240 and 220 kd, respectively. The chain has a single actin-binding domain at its amino terminus. The and chains associate laterally to form dimers, which then join head to head to form tetramers with two actinbinding domains separated by approximately 200 nm. The ends of the spectrin tetramers then associate with short actin filaments, resulting in the spectrin-actin network that forms the cortical cytoskeleton of red blood cells (Figure 11.11). Figure 11.11. Association of the erythrocyte cortical cytoskeleton with the plasma membrane The plasma membrane is associated with a network of spectrin tetramers crosslinked by short actin filaments in association with protein 4.1. The spectrin-actin network is linked to the membrane by ankyrin, which binds to both spectrin and the abundant transmembrane protein band 3. An additional link is provided by the binding of protein 4.1 to glycophorin. The major link between the spectrin-actin network and the plasma membrane is provided by a protein called ankyrin, which binds both to spectrin and to the cytoplasmic domain of an abundant transmembrane protein called band 3. An additional link between the spectrin-actin network and the plasma membrane is provided by protein 4.1, which binds to spectrin-actin junctions as well as recognizing the cytoplasmic domain of glycophorin (another abundant transmembrane protein). Other types of cells contain linkages between the cortical cytoskeleton and the plasma membrane that are similar to those observed in red blood cells. Proteins related to spectrin (nonerythroid spectrin is also called fodrin), ankyrin, and protein 4.1 are expressed in a wide range of cell types, where they fulfill functions analogous to those described for erythrocytes. For example, a family of proteins related to protein 4.1 (the ERM proteins) link actin filaments to the plasma membranes of many different kinds of cells and the spectrin-related protein filamin (see Figure 11.8) constitutes a major link between actin filaments and the plasma membrane of blood platelets. Another member of this group of spectrin-related proteins is dystrophin, which is of particular interest because it is the product of the gene responsible for two types of muscular dystrophy (Duchenne's and Becker's). These X-linked inherited diseases result in progressive degeneration of skeletal muscle, and patients with the more severe form of the disease (Duchenne's muscular dystrophy) usually die in their teens or early twenties. Molecular cloning of the gene responsible for this disorder revealed that it encodes a large protein (427 kd) that is either absent or abnormal in patients with Duchenne's or Becker's muscular dystrophy, respectively. The sequence of dystrophin further indicated that it is related to spectrin, with a single actin-binding domain at its amino terminus and a membranebinding domain at its carboxy terminus. Like spectrin, dystrophin forms dimers that link actin filaments to transmembrane proteins of the muscle cell plasma membrane. These transmembrane proteins in turn link the cytoskeleton to the extracellular matrix, which plays an important role in maintaining cell stability during muscle contraction. In contrast to the uniform surface of red blood cells, most cells have specialized regions of the plasma membrane that form contacts with adjacent cells, tissue components, or other substrates (such as the surface of a culture dish). These regions also serve as attachment sites for bundles of actin filaments that anchor the cytoskeleton to areas of cell contact. These attachments of actin filaments are particularly evident in fibroblasts maintained in tissue culture. Such cultured fibroblasts secrete extracellular matrix proteins that stick to the plastic surface of the culture dish. The fibroblasts then attach to the culture dish via the binding of transmembrane proteins (called integrins) to the extracellular matrix. The sites of attachment are discrete regions (called focal adhesions) that also serve as attachment sites for large bundles of actin filaments called stress fibers. Stress fibers are contractile bundles of actin filaments, crosslinked by -actinin, that anchor the cell and exert tension against the substratum. They are attached to the plasma membrane at focal adhesions via interactions with integrin. These associations, which are complex and not well understood, may be mediated by several other proteins, including talin and vinculin (Figure 11.13). Figure 11.13. Attachment of stress fibers to the plasma membrane at focal adhesions Focal adhesions are mediated by the binding of integrins to proteins of the extracellular matrix. Stress fibers (bundles of actin filaments crosslinked by -actinin) are then bound to the cytoplasmic domain of integrins by complex associations involving a number of proteins. Two possible associations are illustrated: 1) talin binds to both integrin and vinculin, which in turn binds to actin, and 2) integrin binds to -actinin. A number of other proteins (not shown) are also present at focal adhesions and may be involved in anchoring stress fibers to the plasma membrane. For example, both talin and -actinin bind to the cytoplasmic domains of integrins. Talin also binds to vinculin, which in turn interacts with actin. Other proteins found at focal adhesions may also participate in the attachment of actin filaments, and a combination of these interactions may be responsible for the linkage of actin filaments to the plasma membrane. The actin cytoskeleton is similarly anchored to regions of cell-cell contact called adherens junctions (Figure 11.14). In sheets of epithelial cells, these junctions form a continuous beltlike structure (called an adhesion belt) around each cell in which an underlying contractile bundle of actin filaments is linked to the plasma membrane. Contact between cells at adherens junctions is mediated by transmembrane proteins called cadherins. The cadherins form a complex with cytoplasmic proteins called catenins, which associate with actin filaments. Figure 11.14. Attachment of actin filaments to adherens junctions Cell-cell contacts at adherens junctions are mediated by cadherins, which serve as sites of attachment of actin bundles. In sheets of epithelial cells, these junctions form a continuous belt of actin filaments around each cell. The cadherins are transmembrane proteins that bind -catenin to their cytoplasmic domains. -catenin interacts with catenin, which serves as a link to actin filaments. Protrusions of the Cell Surface The surfaces of most cells have a variety of protrusions or extensions that are involved in cell movement, phagocytosis, or specialized functions such as absorption of nutrients. Most of these cell surface extensions are based on actin filaments, which are organized into either relatively permanent or rapidly rearranging bundles or networks. The best-characterized of these actin-based cell surface protrusions are microvilli, fingerlike extensions of the plasma membrane that are particularly abundant on the surfaces of cells involved in absorption, such as the epithelial cells lining the intestine. The microvilli of these cells form a layer on the apical surface (called a brush border) that consists of approximately a thousand microvilli per cell and increases the exposed surface area available for absorption by 10- to 20-fold. In addition to their role in absorption, specialized forms of microvilli, the stereocilia of auditory hair cells, are responsible for hearing by detecting sound vibrations. Their abundance and ease of isolation have facilitated detailed structural analysis of intestinal microvilli, which contain closely packed parallel bundles of 20 to 30 actin filaments (Figure 11.16). Figure 11.16. Organization of microvilli The core actin filaments of microvilli are crosslinked into closely packed bundles by fimbrin and villin. They are attached to the plasma membrane along their length by lateral arms, consisting of myosin I and calmodulin. The plus ends of the actin filaments are embedded in a cap of unidentified proteins at the tip of the microvillus. The filaments in these bundles are crosslinked in part by fimbrin, an actin-bundling protein (discussed earlier) that is present in surface projections of a variety of cell types. However, the major actin-bundling protein in intestinal microvilli is villin, a 95-kd protein present in microvilli of only a few specialized types of cells, such as those lining the intestine and kidney tubules. Along their length, the actin bundles of microvilli are attached to the plasma membrane by lateral arms consisting of the calcium-binding protein calmodulin in association with myosin I, which may be involved in movement of the plasma membrane along the actin bundle of the microvillus. At their base, the actin bundles are anchored in a spectrin-rich region of the actin cortex called the terminal web, which crosslinks and stabilizes the microvilli. In contrast to microvilli, many surface protrusions are transient structures that form in response to environmental stimuli. Several types of these structures extend from the leading edge of a moving cell and are involved in cell locomotion. Pseudopodia are extensions of moderate width, based on actin filaments crosslinked into a threedimensional network, that are responsible for phagocytosis and for the movement of amoebas across a surface. Lamellipodia are broad, sheetlike extensions at the leading edge of fibroblasts, which similarly contain a network of actin filaments. Many cells also extend microspikes or filopodia, thin projections of the plasma membrane supported by actin bundles. The formation and retraction of these structures is based on the regulated assembly and disassembly of actin filaments, as discussed in the following section. Actin, Myosin, and Cell Movement Actin filaments, usually in association with myosin, are responsible for many types of cell movements. Myosin is the prototype of a molecular motor a protein that converts chemical energy in the form of ATP to mechanical energy, thus generating force and movement. The most striking variety of such movement is muscle contraction, which has provided the model for understanding actin-myosin interactions and the motor activity of myosin molecules. However, interactions of actin and myosin are responsible not only for muscle contraction but also for a variety of movements of nonmuscle cells, including cell division, so these interactions play a central role in cell biology. Moreover, the actin cytoskeleton is responsible for the crawling movements of cells across a surface, which appear to be driven directly by actin polymerization as well as actin-myosin interactions. Muscle Contraction Muscle cells are highly specialized for a single task, contraction, and it is this specialization in structure and function that has made muscle the prototype for studying movement at the cellular and molecular levels. There are three distinct types of muscle cells in vertebrates: skeletal muscle, which is responsible for all voluntary movements; cardiac muscle, which pumps blood from the heart; and smooth muscle, which is responsible for involuntary movements of organs such as the stomach, intestine, uterus, and blood vessels. In both skeletal and cardiac muscle, the contractile elements of the cytoskeleton are present in highly organized arrays that give rise to characteristic patterns of cross-striations. It is the characterization of these structures in skeletal muscle that has led to our current understanding of muscle contraction, and other actin-based cell movements, at the molecular level. Skeletal muscles are bundles of muscle fibers, which are single large cells (approximately 50 m in diameter and up to several centimeters in length) formed by the fusion of many individual cells during development (Figure 11.18). Figure 11.18. Structure of muscle cells Muscles are composed of bundles of single large cells (called muscle fibers) that form by cell fusion and contain multiple nuclei. Each muscle fiber contains many myofibrils, which are bundles of actin and myosin filaments organized into a chain of repeating units called sarcomeres. Most of the cytoplasm consists of myofibrils, which are cylindrical bundles of two types of filaments: thick filaments of myosin (about 15 nm in diameter) and thin filaments of actin (about 7 nm in diameter). Each myofibril is organized as a chain of contractile units called sarcomeres, which are responsible for the striated appearance of skeletal and cardiac muscle. The sarcomeres (which are approximately 2.3 m long) consist of several distinct regions, discernible by electron microscopy, which provided critical insights into the mechanism of muscle contraction (Figure 11.19). Figure 11.19. Structure of the sarcomere (B) Diagram showing the organization of actin (thin) and myosin (thick) filaments in the indicated regions. (A, Frank A. Pepe/Biological Photo Service.) The ends of each sarcomere are defined by the Z disc. Within each sarcomere, dark bands (called A bands because they are anisotropic when viewed with polarized light) alternate with light bands (called I bands for isotropic). These bands correspond to the presence or absence of myosin filaments. The I bands contain only thin (actin) filaments, whereas the A bands contain thick (myosin) filaments. The myosin and actin filaments overlap in peripheral regions of the A band, whereas a middle region (called the H zone) contains only myosin. The actin filaments are attached at their plus ends to the Z disc, which includes the crosslinking protein -actinin. The myosin filaments are anchored at the M line in the middle of the sarcomere. Two additional proteins (titin and nebulin) also contribute to sarcomere structure and stability (Figure 11.20). Figure 11.20. Titin and nebulin Molecules of titin extend from the Z disc to the M line and act as springs to keep myosin filaments centered in the sarcomere. Molecules of nebulin extend from the Z disc and are thought to determine the length of associated actin filaments. Titin is an extremely large protein (3000 kd), and single titin molecules extend from the M line to the Z disc. These long molecules of titin are thought to act like springs that keep the myosin filaments centered in the sarcomere and maintain the resting tension that allows a muscle to snap back if overextended. Nebulin filaments are associated with actin and are thought to regulate the assembly of actin filaments by acting as rulers that determine their length. The basis for understanding muscle contraction is the sliding filament model, first proposed in 1954 both by Andrew Huxley and Ralph Niedergerke and by Hugh Huxley and Jean Hanson (Figure 11.21). Figure 11.21. Sliding-filament model of muscle contraction The actin filaments slide past the myosin filaments toward the middle of the sarcomere. The result is shortening of the sarcomere without any change in filament length. During muscle contraction, each sarcomere shortens, bringing the Z discs closer together. There is no change in the width of the A band, but both the I bands and the H zone almost completely disappear. These changes are explained by the actin and myosin filaments sliding past one another, so that the actin filaments move into the A band and H zone. Muscle contraction thus results from an interaction between the actin and myosin filaments that generates their movement relative to one another. The molecular basis for this interaction is the binding of myosin to actin filaments, allowing myosin to function as a motor that drives filament sliding. The type of myosin present in muscle (myosin II) is a very large protein (about 500 kd) consisting of two identical heavy chains (about 200 kd each) and two pairs of light chains (about 20 kd each) (Figure 11.22). Figure 11.22. Myosin II The myosin II molecule consists of two heavy chains and two pairs of light chains (called the essential and regulatory light chains). The heavy chains have globular head regions and long -helical tails, which coil around each other to form dimers. Each heavy chain consists of a globular head region and a long -helical tail. The -helical tails of two heavy chains twist around each other in a coiled-coil structure to form a dimer, and two light chains associate with the neck of each head region to form the complete myosin II molecule. The thick filaments of muscle consist of several hundred myosin molecules, associated in a parallel staggered array by interactions between their tails (Figure 11.23). Figure 11.23. Organization of myosin thick filaments Thick filaments are formed by the association of several hundred myosin II molecules in a staggered array. The globular heads of myosin bind actin, forming cross-bridges between the myosin and actin filaments. The orientation of both actin and myosin filaments reverses at the M line, so their relative polarity is the same on both sides of the sarcomere. The globular heads of myosin bind actin, forming cross-bridges between the thick and thin filaments. It is important to note that the orientation of myosin molecules in the thick filaments reverses at the M line of the sarcomere. The polarity of actin filaments (which are attached to Z discs at their plus ends) similarly reverses at the M line, so the relative orientation of myosin and actin filaments is the same on both halves of the sarcomere. As discussed later, the motor activity of myosin moves its head groups along the actin filament in the direction of the plus end. This movement slides the actin filaments from both sides of the sarcomere toward the M line, shortening the sarcomere and resulting in muscle contraction. In addition to binding actin, the myosin heads bind and hydrolyze ATP, which provides the energy to drive filament sliding. This translation of chemical energy to movement is mediated by changes in the shape of myosin resulting from ATP binding. The generally accepted model (the swinging-cross-bridge model) is that ATP hydrolysis drives repeated cycles of interaction between myosin heads and actin. During each cycle, conformational changes in myosin result in the movement of myosin heads along actin filaments. Although the molecular mechanisms are still not fully understood, a plausible working model for myosin function has been derived both from in vitro studies of myosin movement along actin filaments (a system developed by James Spudich and Michael Sheetz) and from determination of the three-dimensional structure of myosin by Ivan Rayment and his colleagues (Figure 11.24). Figure 11.24. Model for myosin action The binding of ATP dissociates myosin from actin. ATP hydrolysis then induces a conformational change that displaces the myosin head group. This is followed by binding of the myosin head to a new position on the actin filament and release of ADP and P i. The return of the myosin head to its original conformation drives actin filament sliding. The cycle starts with myosin (in the absence of ATP) tightly bound to actin. ATP binding dissociates the myosin-actin complex and the hydrolysis of ATP then induces a conformational change in myosin. This change affects the neck region of myosin that binds the light chains (see Figure 11.22), which acts as a lever arm to displace the myosin head by about 5 nm. The products of hydrolysis (ADP and Pi) remain bound to the myosin head, which is said to be in the "cocked" position. The myosin head then rebinds at a new position on the actin filament, resulting in the release of ADP and Pi and triggering the "power stroke," in which the myosin head returns to its initial conformation, thereby sliding the actin filament toward the M line of the sarcomere. The contraction of skeletal muscle is triggered by nerve impulses, which stimulate the release of Ca 2+ from the sarcoplasmic reticulum a specialized network of internal membranes, similar to the endoplasmic reticulum, that stores high concentrations of Ca2+ ions. The release of Ca2+ from the sarcoplasmic reticulum increases the concentration of Ca2+ in the cytosol from approximately 10-7 to 10-5M. The increased Ca2+ concentration signals muscle contraction via the action of two accessory proteins bound to the actin filaments: tropomyosin and troponin (Figure 11.25). Figure 11.25. Association of tropomyosin and troponins with actin filaments (A) Tropomyosin binds lengthwise along actin filaments and, in striated muscle, is associated with a complex of three troponins: troponin I (TnI), troponin C (TnC), and troponin T (TnT). In the absence of Ca2+, the tropomyosin-troponin complex blocks the binding of myosin to actin. Binding of Ca 2+ to TnC shifts the complex, relieving this inhibition and allowing contraction to proceed. (B) Cross-sectional view. Tropomyosin is a fibrous protein that binds lengthwise along the groove of actin filaments. In striated muscle, each tropomyosin molecule is bound to troponin, which is a complex of three polypeptides: troponin C (Ca2+-binding), troponin I (inhibitory), and troponin T (tropomyosin-binding). When the concentration of Ca2+ is low, the complex of the troponins with tropomyosin blocks the interaction of actin and myosin, so the muscle does not contract. At high concentrations, Ca2+ binding to troponin C shifts the position of the complex, relieving this inhibition and allowing contraction to proceed. Contractile Assemblies of Actin and Myosin in Nonmuscle Cells Contractile assemblies of actin and myosin, resembling smallscale versions of muscle fibers, are present also in nonmuscle cells. As in muscle, the actin filaments in these contractile assemblies are interdigitated with bipolar filaments of myosin II, consisting of 15 to 20 myosin II molecules, which produce contraction by sliding the actin filaments relative to one another (Figure 11.26). The actin filaments in contractile bundles in nonmuscle cells are also associated with tropomyosin, which facilitates their interaction with myosin II, probably by competing with filamin for binding sites on actin. Figure 11.26. Contractile assemblies in nonmuscle cells Bipolar filaments of myosin II produce contraction by sliding actin filaments in opposite directions. Two examples of contractile assemblies in nonmuscle cells, stress fibers and adhesion belts, were discussed earlier with respect to attachment of the actin cytoskeleton to regions of cell-substrate and cell-cell contacts (see Figures 11.13 and 11.14). The contraction of stress fibers produces tension across the cell, allowing the cell to pull on a substrate (e.g., the extracellular matrix) to which it is anchored. The contraction of adhesion belts alters the shape of epithelial cell sheets: a process that is particularly important during embryonic development, when sheets of epithelial cells fold into structures such as tubes. The most dramatic example of actin-myosin contraction in nonmuscle cells, however, is provided by cytokinesis the division of a cell into two following mitosis (Figure 11.27). Figure 11.27. Cytokinesis Following completion of mitosis (nuclear division), a contractile ring consisting of actin filaments and myosin II divides the cell in two. Toward the end of mitosis in animal cells, a contractile ring consisting of actin filaments and myosin II assembles just underneath the plasma membrane. Its contraction pulls the plasma membrane progressively inward, constricting the center of the cell and pinching it in two. Interestingly, the thickness of the contractile ring remains constant as it contracts, implying that actin filaments disassemble as contraction proceeds. The ring then disperses completely following cell division. The regulation of actin-myosin contraction in striated muscle, discussed earlier, is mediated by the binding of Ca2+ to troponin. In nonmuscle cells and in smooth muscle, however, contraction is regulated primarily by phosphorylation of one of the myosin light chains, called the regulatory light chain (Figure 11.28). Figure 11.28. Regulation of myosin by phosphorylation Ca2+ binds to calmodulin, which in turn binds to myosin light-chain kinase (MLCK). The active calmodulin-MLCK complex then phosphorylates the myosin II regulatory light chain, converting myosin from an inactive to an active state. Phosphorylation of the regulatory light chain in these cells has at least two effects: It promotes the assembly of myosin into filaments, and it increases myosin catalytic activity, enabling contraction to proceed. The enzyme that catalyzes this phosphorylation, called myosin light-chain kinase, is itself regulated by association with the Ca2+binding protein calmodulin. Increases in cytosolic Ca2+ promote the binding of calmodulin to the kinase, resulting in phosphorylation of the myosin regulatory light chain. Increases in cytosolic Ca2+ are thus responsible, albeit indirectly, for activating myosin in smooth muscle and nonmuscle cells, as well as in striated muscle. Unconventional Myosins In addition to myosin II ("conventional" two-headed myosin), several other types of myosin are found in nonmuscle cells. In contrast to myosin II, these "unconventional" myosins do not form filaments and therefore are not involved in contraction. They may, however, be involved in a variety of other kinds of cell movements, such as the transport of membrane vesicles and organelles along actin filaments, phagocytosis, and extension of pseudopods in amoebae. Figure 11.29. Myosin I Myosin I contains a head group similar to myosin II, but it has a comparatively short tail and does not form dimers or filaments. Although it cannot induce contraction, myosin I can move along actin filaments (toward the plus end), carrying a variety of cargoes (such as membrane vesicles) attached to its tail. The best-studied of these unconventional myosins are members of the myosin I family (Figure 11.29). The myosin I proteins contain a globular head group that acts as a molecular motor, like that of myosin II. However, members of the myosin I family are much smaller molecules (about 110 kd in mammalian cells) that lack the long tail of myosin II and do not form dimers. Their tails can instead bind to other structures, such as membrane vesicles or organelles. The movement of myosin I along an actin filament can then transport its attached cargo. One function of myosin I, discussed earlier, is to form the lateral arms that link actin bundles to the plasma membrane of intestinal microvilli (see Figure 11.16). In these structures, the motor activity of myosin I may move the plasma membrane along the actin bundles, toward the tip of the microvillus. Additional functions of myosin I may be in the transport of vesicles and organelles along actin filaments and in movement of the plasma membrane during phagocytosis and pseudopod extension. In addition to myosins I and II, at least 12 other classes of unconventional myosins (III through XIV) have been identified. Some of these unconventional myosins are two-headed like myosin II, whereas others are one-headed like myosin I. The functions of most of these unconventional myosins remain to be determined, but some have been clearly shown to play important roles in organelle movement (myosins V and VI) and in sensory functions such as vision (myosin III) and hearing (myosins VI and VII). Cell Crawling The crawling movements of cells across a surface represent a basic form of cell locomotion, employed by a wide variety of different kinds of cells. Examples include the movements of amoebas, the migration of embryonic cells during development, the invasion of tissues by white blood cells to fight infection, the migration of cells involved in wound healing, and the spread of cancer cells during the metastasis of malignant tumors. Similar types of movement are also responsible for phagocytosis and for the extension of nerve cell processes during development of the nervous system. All of these movements are based on the dynamic properties of the actin cytoskeleton, although the detailed mechanisms involved remain to be fully understood. Cell crawling involves a coordinated cycle of movements, which can be viewed in three stages. First, protrusions such as pseudopodia, lamellipodia, or microspikes must be extended from the leading edge of the cell (Figure 11.30). Second, these extensions must attach to the substratum across which the cell is migrating. Finally, the trailing edge of the cell must dissociate from the substratum and retract into the cell body. Figure 11.30. Cell crawling The crawling movements of cells across a surface can be viewed as three stages of coordinated movements: (1) extension of the leading edge, (2) attachment of the leading edge to the substratum, and (3) retraction of the rear of the cell into the cell body A variety of experiments indicate that extension of the leading edge involves the polymerization and crosslinking of actin filaments. For example, inhibition of actin polymerization (e.g., by treatment with cytochalasin) blocks the formation of cell surface protrusions. The regulated turnover of actin filaments, as illustrated in Figure 11.5, leads to the extension of processes such as filopodia and lamellipodia at the leading edge of the cell, and both cofilin and Arp2/3 proteins appear to be involved in this process. Unconventional myosins may also participate in the extension of processes at the leading edge: Myosin I is required for pseudopod extension in the amoeba Dictyostelium and Myosin V for extension of filopodia in neurons. Following their extension, protrusions from the leading edge must attach to the substratum in order to function in cell locomotion. For slow-moving cells, such as fibroblasts, attachment involves the formation of focal adhesions (see Figure 11.13). Cells moving more rapidly, such as amoebas or white blood cells, form more diffuse contacts with the substratum, the molecular composition of which is not known. The third stage of cell crawling, retraction of the trailing edge, is the least understood. The attachments of the trailing edge to the substratum are broken, and the rear of the cell recoils into the cell body. The process appears to require the development of tension between the front and rear of the cell, generating contractile force that eventually pulls the rear of the cell forward. This aspect of cell locomotion is impaired in mutants of Dictyostelium lacking myosin II, consistent with a role for myosin II in contracting the actin cortex and generating the force required for retraction of the trailing edge. Intermediate Filaments Intermediate filaments have a diameter of about 10 nm, which is intermediate between the diameters of the two other principal elements of the cytoskeleton, actin filaments (about 7 nm) and microtubules (about 25 nm). In contrast to actin filaments and microtubules, the intermediate filaments are not directly involved in cell movements. Instead, they appear to play basically a structural role by providing mechanical strength to cells and tissues. Intermediate Filament Proteins Whereas actin filaments and microtubules are polymers of single types of proteins (actin and tubulin, respectively), intermediate filaments are composed of a variety of proteins that are expressed in different types of cells. More than 50 different intermediate filament proteins have been identified and classified into six groups based on similarities between their amino acid sequences (Table 11.1). Table 11.1. Intermediate Filament Proteins Type I II III IV V VI Protein Size (kd) Site of expression Acidic keratins (~15 proteins) Neutral or basic keratins (~15 proteins) Vimentin Desmin Glial fibrillary acidic protein Peripherin Neurofilament proteins NF-L NF-M NF-H 40 60 Epithelial cells 50 70 Epithelial cells 54 53 51 57 Fibroblasts, white blood cells, and other cell types Muscle cells Glial cells Peripheral neurons -Internexin 67 150 200 66 Neurons Neurons Neurons Neurons Nuclear lamins Nestin 60 75 200 Nuclear lamina of all cell types Stem cells of central nervous system Types I and II consist of two groups of keratins, each consisting of about 15 different proteins, which are expressed in epithelial cells. Each type of epithelial cell synthesizes at least one type I (acidic) and one type II (neutral/basic) keratin, which copolymerize to form filaments. Some type I and II keratins (called hard keratins) are used for production of structures such as hair, nails, and horns. The other type I and II keratins (soft keratins) are abundant in the cytoplasm of epithelial cells, with different keratins being expressed in various differentiated cell types. The type III intermediate filament proteins include vimentin, which is found in a variety of different kinds of cells, including fibroblasts, smooth muscle cells, and white blood cells. Another type III protein, desmin, is specifically expressed in muscle cells, where it connects the Z discs of individual contractile elements. A third type III intermediate filament protein is specifically expressed in glial cells, and a fourth is expressed in neurons of the peripheral nervous system. The type IV intermediate filament proteins include the three neurofilament (NF) proteins (designated NF-L, NFM, and NF-H for light, medium, and heavy, respectively). These proteins form the major intermediate filaments of many types of mature neurons. They are particularly abundant in the axons of motor neurons and are thought to play a critical role in supporting these long, thin processes, which can extend more than a meter in length. Another type IV protein (-internexin) is expressed at an earlier stage of neuron development, prior to expression of the neurofilament proteins. The single type VI intermediate filament protein (nestin) is expressed even earlier during the development of neurons, in stem cells of the central nervous system. The type V intermediate filament proteins are the nuclear lamins, which are found in most eukaryotic cells. Rather than being part of the cytoskeleton, the nuclear lamins are components of the nuclear envelope. They also differ from the other intermediate filament proteins in that they assemble to form an orthogonal meshwork underlying the nuclear membrane. Despite considerable diversity in size and amino acid sequence, the various intermediate filament proteins share a common structural organization (Figure 11.31). Figure 11.31. Structure of intermediate filament proteins Intermediate filament proteins contain a central -helical rod domain of approximately 310 amino acids (350 amino acids in the nuclear lamins). The N-terminal head and C-terminal tail domains vary in size and shape. All of the intermediate filament proteins have a central -helical rod domain of approximately 310 amino acids (350 amino acids in the nuclear lamins). This central rod domain is flanked by amino- and carboxy-terminal domains, which vary among the different intermediate filament proteins in size, sequence, and secondary structure. As discussed next, the -helical rod domain plays a central role in filament assembly, while the variable head and tail domains presumably determine the specific functions of the different intermediate filament proteins. Assembly of Intermediate Filaments The first stage of filament assembly is the formation of dimers in which the central rod domains of two polypeptide chains are wound around each other in a coiled-coil structure, similar to that formed by myosin II heavy chains (Figure 11.32). Figure 11.32. Assembly of intermediate filaments The central rod domains of two polypeptides wind around each other in a coiled-coil structure to form dimers. Dimers then associate in a staggered antiparallel fashion to form tetramers. Tetramers associate end to end to form protofilaments and laterally to form filaments. Each filament contains approximately eight protofilaments wound around each other in a ropelike structure. The dimers then associate in a staggered antiparallel fashion to form tetramers, which can assemble end to end to form protofilaments. The final intermediate filament contains approximately eight protofilaments wound around each other in a ropelike structure. Because they are assembled from antiparallel tetramers, both ends of intermediate filaments are equivalent. Consequently, in contrast to actin filaments and microtubules, intermediate filaments are apolar; they do not have distinct plus and minus ends. Filament assembly requires interactions between specific types of intermediate filament proteins. For example, keratin filaments are always assembled from heterodimers containing one type I and one type II polypeptide. In contrast, the type III proteins can assemble into filaments containing only a single polypeptide (e.g., vimentin) or consisting of two different type III proteins (e.g., vimentin plus desmin). The type III proteins do not, however, form copolymers with the keratins. Among the type IV proteins, -internexin can assemble into filaments by itself, whereas the three neurofilament proteins copolymerize to form heteropolymers. Intermediate filaments are generally more stable than actin filaments or microtubules and do not exhibit the dynamic behavior associated with these other elements of the cytoskeleton (e.g., the treadmilling of actin filaments illustrated in Figure 11.4). However, intermediate filament proteins are frequently modified by phosphorylation, which can regulate their assembly and disassembly within the cell. The clearest example is phosphorylation of the nuclear lamins (see Figure 8.31), which results in disassembly of the nuclear lamina and breakdown of the nuclear envelope during mitosis. Cytoplasmic intermediate filaments, such as vimentin, are also phosphorylated at mitosis, which can lead to their disassembly and reorganization in dividing cells. Figure 8.31. Dissolution of the nuclear lamina The nuclear lamina consists of a meshwork of lamin filaments. At mitosis, Cdc2 and other protein kinases phosphorylate the lamins, causing the filaments to dissociate into free lamin dimers. Intracellular Organization of Intermediate Filaments Intermediate filaments form an elaborate network in the cytoplasm of most cells, extending from a ring surrounding the nucleus to the plasma membrane. Both keratin and vimentin filaments attach to the nuclear envelope, apparently serving to position and anchor the nucleus within the cell. In addition, intermediate filaments can associate not only with the plasma membrane but also with the other elements of the cytoskeleton, actin filaments and microtubules. Intermediate filaments thus provide a scaffold that integrates the components of the cytoskeleton and organizes the internal structure of the cell. The keratin filaments of epithelial cells are tightly anchored to the plasma membrane at two areas of specialized cell contacts, desmosomes and hemidesmosomes (Figure 11.34). Figure 11.34. Attachment of intermediate filaments to desmosomes and hemidesmosomes (B) Schematic of a desmosome. Intermediate filaments are anchored to sites of cell-cell adhesion by desmoplaskin. (C) Schematic of a hemidesmosome. Intermediate filaments are anchored to an integrin by plectin. Desmosomes are junctions between adjacent cells, at which cell-cell contacts are mediated by transmembrane proteins related to the cadherins. On their cytoplasmic side, desmosomes are associated with a characteristic dense plaque of intracellular proteins, to which keratin filaments are attached. These attachments are mediated by desmoplakin, a member of a family of proteins called plakins that bind intermediate filaments and link them to other cellular structures. Hemidesmosomes are morphologically similar junctions between epithelial cells and underlying connective tissue, at which keratin filaments are linked by different members of the plakin family (e.g., plectin) to integrins. Desmosomes and hemidesmosomes thus anchor intermediate filaments to regions of cell-cell and cell-substratum contact, respectively, similar to the attachment of the actin cytoskeleton to the plasma membrane at adherens junctions and focal adhesions. It is important to note that the keratin filaments anchored to both sides of desmosomes serve as a mechanical link between adjacent cells in an epithelial layer, thereby providing mechanical stability to the entire tissue. In addition to linking intermediate filaments to cell junctions, some plakins link intermediate filaments to other elements of the cytoskeleton. Plectin, for example, binds actin filaments and microtubules in addition to intermediate filaments, so it can provide bridges between these cytoskeletal components. These bridges to intermediate filaments are thought to brace and stabilize actin filaments and microtubules, thereby increasing the mechanical stability of the cell. Two types of intermediate filaments, desmin and the neurofilaments, play specialized roles in muscle and nerve cells, respectively. Desmin connects the individual actin-myosin assemblies of muscle cells both to one another and to the plasma membrane, thereby linking the actions of individual contractile elements. Neurofilaments are the major intermediate filaments in most mature neurons. They are particularly abundant in the long axons of motor neurons, where they appear to be anchored to actin filaments and microtubules by neuronal members of the plakin family. Neurofilaments are thought to play an important role in providing mechanical support and stabilizing other elements of the cytoskeleton in these long, thin extensions of nerve cells. Functions of Keratins and Neurofilaments: Diseases of the Skin and Nervous System Although intermediate filaments have long been thought to provide structural support to the cell, direct evidence for their function has only recently been obtained. Some cells in culture make no intermediate filament proteins, indicating that these proteins are not required for the growth of cells in vitro. Similarly, injection of cultured cells with antibody against vimentin disrupts intermediate filament networks without affecting cell growth or movement. Therefore, it has been thought that intermediate filaments are most needed to strengthen the cytoskeleton of cells in the tissues of multicellular organisms, where they are subjected to a variety of mechanical stresses that do not affect cells in the isolated environment of a culture dish. Experimental evidence for such an in vivo role of intermediate filaments was first provided in 1991 by studies in the laboratory of Elaine Fuchs. These investigators used transgenic mice to investigate the in vivo effects of expressing a keratin deletion mutant encoding a truncated polypeptide that disrupted the formation of normal keratin filaments (Figure 11.36). This mutant keratin gene was introduced into transgenic mice, where it was expressed in basal cells of the epidermis and disrupted formation of a normal keratin cytoskeleton. This resulted in the development of severe skin abnormalities, including blisters due to epidermal cell lysis following mild mechanical trauma, such as rubbing of the skin. The skin abnormalities of these transgenic mice thus provided direct support for the presumed role of keratins in providing mechanical strength to epithelial cells in tissues. Figure 11.36. Experimental demonstration of keratin function A plasmid encoding a mutant keratin that interferes with the normal assembly of keratin filaments was microinjected into one pronucleus of a fertilized egg. Microinjected embryos were then transferred to a foster mother, and some of the offspring were found to have incorporated the mutant keratin gene into their genome. Expression of the mutant gene in these transgenic mice disrupted the keratin cytoskeleton of cells of the epidermis, resulting in severe skin blistering due to cell lysis following mild mechanical stress. These experiments also pointed to the molecular basis of a human genetic disease, epidermolysis bullosa simplex (EBS). Like the transgenic mice expressing mutant keratin genes, patients with this disease develop skin blisters resulting from cell lysis after minor trauma. This similarity prompted studies of the keratin genes in EBS patients, leading to the demonstration that EBS is caused by keratin gene mutations that interfere with the normal assembly of keratin filaments. Thus, both experimental studies in transgenic mice and molecular analysis of a human genetic disease have demonstrated the role of keratins in allowing skin cells to withstand mechanical stress. Continuing studies have shown that mutations in other keratins are responsible for several other inherited skin diseases, which are similarly characterized by abnormal fragility of epidermal cells. Other studies in transgenic mice have implicated abnormalities of neurofilaments in diseases of motor neurons, particularly amyotrophic lateral sclerosis (ALS). ALS, known as Lou Gehrig's disease and the disease afflicting the renowned physicist Stephen Hawking, results from progressive loss of motor neurons, which in turn leads to muscle atrophy, paralysis, and eventual death. ALS and other types of motor neuron disease are characterized by the accumulation and abnormal assembly of neurofilaments, suggesting that neurofilament abnormalities might contribute to these pathologies. Consistent with this possibility, overexpression of NF-L or NF-H in transgenic mice has been found to result in the development of a condition similar to ALS. Although the mechanism involved remains to be understood, these experiments clearly suggest the involvement of neurofilaments in the pathogenesis of motor neuron disease. Microtubules Microtubules, the third principal component of the cytoskeleton, are rigid hollow rods approximately 25 nm in diameter. Like actin filaments, microtubules are dynamic structures that undergo continual assembly and disassembly within the cell. They function both to determine cell shape and in a variety of cell movements, including some forms of cell locomotion, the intracellular transport of organelles, and the separation of chromosomes during mitosis. Structure, Assembly, and Dynamic Instability of Microtubules In contrast to intermediate filaments, which are composed of a variety of different fibrous proteins, microtubules are composed of a single type of globular protein, called tubulin. Tubulin is a dimer consisting of two closely related 55-kd polypeptides, -tubulin and -tubulin. Like actin, both - and -tubulin are encoded by small families of related genes. In addition, a third type of tubulin (-tubulin) is specifically localized to the centrosome, where it plays a critical role in initiating microtubule assembly (discussed shortly). Tubulin dimers polymerize to form microtubules, which generally consist of 13 linear protofilaments assembled around a hollow core (Figure 11.37). Figure 11.37. Structure of microtubules Dimers of - and -tubulin polymerize to form microtubules, which are composed of 13 protofilaments assembled around a hollow core The protofilaments, which are composed of head-to-tail arrays of tubulin dimers, are arranged in parallel. Consequently, microtubules (like actin filaments) are polar structures with two distinct ends: a fast-growing plus end and a slow-growing minus end. This polarity is an important consideration in determining the direction of movement along microtubules, just as the polarity of actin filaments defines the direction of myosin movement. Tubulin dimers can depolymerize as well as polymerize, and microtubules can undergo rapid cycles of assembly and disassembly. Both - and -tubulin bind GTP, which functions analogously to the ATP bound to actin to regulate polymerization. In particular, the GTP bound to -tubulin (though not that bound to -tubulin) is hydrolyzed to GDP during or shortly after polymerization. This GTP hydrolysis weakens the binding affinity of tubulin for adjacent molecules, thereby favoring depolymerization and resulting in the dynamic behavior of microtubules. Like actin filaments (see Figure 11.4), microtubules undergo treadmilling, a dynamic behavior in which tubulin molecules bound to GDP are continually lost from the minus end and replaced by the addition of tubulin molecules bound to GTP to the plus end of the same microtubule. In microtubules, GTP hydrolysis also results in the behavior known as dynamic instability, in which individual microtubules alternate between cycles of growth and shrinkage (Figure 11.38). Figure 11.38. Dynamic instability of microtubules Dynamic instability results from the hydrolysis of GTP bound to -tubulin during or shortly after polymerization, which reduces its binding affinity for adjacent molecules. Growth of microtubules continues as long as there is a high concentration of tubulin bound to GTP. New GTP-bound tubulin molecules are then added more rapidly than GTP is hydrolyzed, so a GTP cap is retained at the growing end. However, if GTP is hydrolyzed more rapidly than new subunits are then added, the presence of GDP-bound tubulin at the end of the microtubule leads to disassembly and shrinkage. Only the plus ends of microtubules are illustrated. Whether a microtubule grows or shrinks is determined by the rate of tubulin addition relative to the rate of GTP hydrolysis. As long as new GTP-bound tubulin molecules are added more rapidly than GTP is hydrolyzed, the microtubule retains a GTP cap at its plus end and microtubule growth continues. However, if the rate of polymerization slows, the GTP bound to tubulin at the plus end of the microtubule will be hydrolyzed to GDP. If this occurs, the GDP-bound tubulin will dissociate, resulting in rapid depolymerization and shrinkage of the microtubule. Dynamic instability, described by Tim Mitchison and Marc Kirschner in 1984, results in the continual and rapid turnover of most microtubules, which have half-lives of only several minutes within the cell. As discussed later, this rapid turnover of microtubules is particularly critical for the remodeling of the cytoskeleton that occurs during mitosis. Because of the central role of microtubules in mitosis, drugs that affect microtubule assembly are useful not only as experimental tools in cell biology but also in the treatment of cancer. Colchicine and colcemid are examples of commonly used experimental drugs that bind tubulin and inhibit microtubule polymerization, which in turn blocks mitosis. Two related drugs (vincristine and vinblastine) are used in cancer chemotherapy because they selectively inhibit rapidly dividing cells. Another useful drug, taxol, stabilizes microtubules rather than inhibiting their assembly. Such stabilization also blocks cell division, and taxol is used as an anticancer agent as well as an experimental tool. The Centrosome and Microtubule Organization The microtubules in most cells extend outward from a microtubule-organizing center, in which the minus ends of microtubules are anchored. In animal cells, the major microtubule-organizing center is the centrosome, which is located adjacent to the nucleus near the center of interphase (nondividing) cells (Figure 11.39). Figure 11.39. Intracellular organization of microtubules The minus ends of microtubules are anchored in the centrosome. In interphase cells, the centrosome is located near the nucleus and microtubules extend outward to the cell periphery. During mitosis, duplicated centrosomes separate and microtubules reorganize to form the mitotic spindle. During mitosis, microtubules similarly extend outward from duplicated centrosomes to form the mitotic spindle, which is responsible for the separation and distribution of chromosomes to daughter cells. The centrosome thus plays a key role in determining the intracellular organization of microtubules, although most details of its function remain a mystery. The centrosome serves as the initiation site for the assembly of microtubules, which grow outward from the centrosome toward the periphery of the cell. This can be clearly visualized in cells that have been treated with colcemid to disassemble their microtubules. When the drug is removed, the cells recover and new microtubules can be seen growing outward from the centrosome. Importantly, the initiation of microtubule growth at the centrosome establishes the polarity of microtubules within the cell. In particular, microtubules grow by the addition of tubulin to their plus ends, which extend outward from the centrosome toward the cell periphery. The centrosomes of most animal cells contain a pair of centrioles, oriented perpendicular to each other, surrounded by amorphous pericentriolar material. The centrioles are cylindrical structures consisting of nine triplets of microtubules, similar to the basal bodies of cilia and flagella (discussed later in the chapter). Although centrioles are probably the precursors of basal bodies, they appear to be dispensible for the function of the centrosome. Centrioles do not appear to be required for the assembly or organization of microtubules, and they are not found in plant cells, many unicellular eukaryotes, and some animal cells (such as mouse eggs). The microtubules that emanate from the centrosome terminate in the pericentriolar material, not the centrioles, and it is the pericentriolar material that initiates microtubule assembly. The key protein in the centrosome that nucleates assembly of microtubules is -tubulin, a minor species of tubulin first identified in fungi. Complexes of -tubulin form ring structures that contain 10 to 13 -tubulin molecules and have diameters similar to those of microtubules. These -tubulin rings serve as nucleation sites for the assembly of microtubules and may remain bound to their minus ends. Reorganization of Microtubules during Mitosis As noted earlier, microtubules completely reorganize during mitosis, providing a dramatic example of the importance of their dynamic instability. The microtubule array present in interphase cells disassembles and the free tubulin subunits are reassembled to form the mitotic spindle, which is responsible for the separation of daughter chromosomes. This restructuring of the microtubule cytoskeleton is directed by duplication of the centrosome to form two separate microtubule-organizing centers at opposite poles of the mitotic spindle. The centrioles and other components of the centrosome are duplicated in interphase cells, but they remain together on one side of the nucleus until the beginning of mitosis (Figure 11.43). Figure 11.43. Formation of the mitotic spindle The centrioles and centrosomes duplicate during interphase. During prophase of mitosis, the duplicated centrosomes separate and move to opposite sides of the nucleus. The nuclear envelope then disassembles, and microtubules reorganize to form the mitotic spindle. Kinetochore microtubules are attached to the condensed chromosomes, polar microtubules overlap with each other in the center of the cell, and astral microtubules extend outward to the cell periphery. At metaphase, the condensed chromosomes are aligned at the center of the spindle. The two centrosomes then separate and move to opposite sides of the nucleus, forming the two poles of the mitotic spindle. As the cell enters mitosis, the dynamics of microtubule assembly and disassembly also change dramatically. First, the rate of microtubule disassembly increases about tenfold, resulting in overall depolymerization and shrinkage of microtubules. At the same time, the number of microtubules emanating from the centrosome increases by five- to tenfold. In combination, these changes result in disassembly of the interphase microtubules and the outgrowth of large numbers of short microtubules from the centrosomes. As first proposed by Marc Kirschner and Tim Mitchison in 1986, formation of the mitotic spindle involves the selective stabilization of some of the microtubules radiating from the centrosomes. These microtubules are of three types, two of which make up the mitotic spindle. Kinetochore microtubules attach to the condensed chromosomes of mitotic cells at their centromeres, which are associated with specific proteins to form the kinetochore (see Figure 4.16). Attachment to the kinetochore stabilizes these microtubules, which, as discussed below, play a critical role in separation of the mitotic chromosomes. The second type of microtubules found in the mitotic spindle (polar microtubules) are not attached to chromosomes. Instead, the polar microtubules emanating from the two centrosomes are stabilized by overlapping with each other in the center of the cell. Astral microtubules extend outward from the centrosomes to the cell periphery and have freely exposed plus ends. As discussed later, both the polar and astral microtubules also contribute to chromosome movement by pushing the spindle poles apart. Figure 4.16. The centromere of a metaphase chromosome The centromere is the region at which the two sister chromatids remain attached at metaphase. Specific proteins bind to centromeric DNA, forming the kinetochore, which is the site of spindle fiber attachment. As mitosis proceeds, the condensed chromosomes first align on the metaphase plate and then separate, with the two chromatids of each chromosome being pulled to opposite poles of the spindle. Chromosome movement is mediated by motor proteins associated with the spindle microtubules, as will be discussed shortly. In the final stage of mitosis, nuclear envelopes re-form, the chromosomes decondense, and cytokinesis takes place. Each daughter cell then contains one centrosome, which nucleates the formation of a new network of interphase microtubules. Stabilization of Microtubules and Cell Polarity Because of their inherent dynamic instability, most microtubules are frequently disassembled within the cell. This dynamic behavior can, however, be modified by the interactions of microtubules with other proteins. Some cellular proteins act to disassemble microtubules, either by severing microtubules or by increasing the rate of tubulin depolymerization from microtubule ends. Other proteins (called microtubule-associated proteins or MAPs) bind to microtubules and increase their stability. Such interactions allow the cell to stabilize microtubules in particular locations and provide an important mechanism for determining cell shape and polarity. A large number of MAPs have been identified, and they vary depending on the type of cell. The best-characterized are MAP-1, MAP-2, and tau, isolated from neuronal cells, and MAP-4, which is present in all non-neuronal vertebrate cell types. The tau protein has been extensively studied because it is the main component of the characteristic lesions found in the brains of Alzheimer patients. The activity of MAPs is regulated by phosphorylation, allowing the cell to control microtubule stability. A good example of the role of stable microtubules in determining cell polarity is provided by nerve cells, which consist of two distinct types of processes (axons and dendrites) extending from a cell body (Figure 11.44). Figure 11.44. Organization of microtubules in nerve cells Two distinct types of processes extend from the cell body of nerve cells (neurons). Dendrites are short processes that receive stimuli from other nerve cells. The single long axon then carries impulses from the cell body to other cells, which may be either other neurons or an effector cell, such as a muscle. Stable microtubules in both axons and dendrites terminate in the cytoplasm rather than being anchored in the centrosome. In dendrites, microtubules are oriented in both directions, with their plus ends pointing both toward and away from the cell body. In contrast, all of the axon microtubules are oriented with their plus ends pointing toward the tip of the axon. Both axons and dendrites are supported by stable microtubules, together with the neurofilaments discussed in the preceding section of this chapter. However, the microtubules in axons and dendrites are organized differently and associated with distinct MAPs. In axons, the microtubules are all oriented with their plus ends away from the cell body, similar to the general orientation of microtubules in other cell types. The minus ends of most of the microtubules in axons, however, are not anchored in the centrosome; instead, both the plus and minus ends of these microtubules terminate in the cytoplasm of the axon. In dendrites, the microtubules are oriented in both directions; some plus ends point toward the cell body and some point toward the cell periphery. These distinct microtubule arrangements are paralleled by differences in MAPs: Axons contain tau proteins, but no MAP-2, whereas dendrites contain MAP-2, but no tau proteins, and it appears that these differences in MAP-2 and tau distribution are responsible for the distinct organization of stable microtubules in axons and dendrites. Microtubule Motors and Movements Microtubules are responsible for a variety of cell movements, including the intracellular transport and positioning of membrane vesicles and organelles, the separation of chromosomes at mitosis, and the beating of cilia and flagella. As discussed for actin filaments earlier in this chapter, movement along microtubules is based on the action of motor proteins that utilize energy derived from ATP hydrolysis to produce force and movement. Members of two large families of motor proteins the kinesins and the dyneins are responsible for powering the variety of movements in which microtubules participate. Identification of Microtubule Motor Proteins Kinesin and dynein, the prototypes of microtubule motor proteins, move along microtubules in opposite directions kinesin toward the plus end and dynein toward the minus end (Figure 11.45). Figure 11.45. Microtubule motor proteins Kinesin and dynein move in opposite directions along microtubules, toward the plus and minus ends, respectively. Kinesin consists of two heavy chains, wound around each other in a coiled-coil structure, and two light chains. The globular head domains of the heavy chains bind microtubules and are the motor domains of the molecule. Dynein consists of two or three heavy chains (two are shown here) in association with multiple light and intermediate chains. The globular head domains of the heavy chains are the motor domains. The first of these microtubule motor proteins to be identified was dynein, which was isolated by Ian Gibbons in 1965. The purification of this form of dynein (called axonemal dynein) was facilitated because it is a highly abundant protein in cilia, just as the abundance of myosin facilitated its isolation from muscle cells. The identification of other microtubule-based motors, however, was more problematic because the proteins responsible for processes such as chromosome movement and organelle transport are present at comparatively low concentrations in the cytoplasm. Isolation of these proteins therefore depended on the development of new experimental methods to detect the activity of molecular motors in cell-free systems. The development of in vitro assays for cytoplasmic motor proteins was based on the use of video-enhanced microscopy, developed by Robert Allen and Shinya Inoué in the early 1980s, to study the movement of membrane vesicles and organelles along microtubules in squid axons. In this method, a video camera is used to increase the contrast of images obtained with the light microscope, substantially improving the detection of small objects and allowing the movement of organelles to be followed in living cells. Using this approach, Allen, Scott Brady, and Ray Lasek demonstrated that organelle movements also took place in a cell-free system in which the plasma membrane had been removed and a cytoplasmic extract had been spread on a glass slide. These observations led to the development of an in vitro reconstructed system, which provided an assay capable of detecting cellular proteins responsible for organelle movement. In 1985 Brady, as well as Ronald Vale, Thomas Reese, and Michael Sheetz, capitalized on these developments to identify kinesin as a novel microtubule motor protein, present in both squid axons and bovine brain. Further studies demonstrated that kinesin translocates along microtubules in only a single direction toward the plus end. Because the plus ends of microtubules in axons are all oriented away from the cell body (see Figure 11.44), the movement of kinesin in this direction transports vesicles and organelles away from the cell body, toward the tip of the axon. Within intact axons, however, vesicles and organelles also had been observed to move back toward the cell body, implying that a different motor protein might be responsible for movement along microtubules in the opposite direction toward the minus end. Consistent with this prediction, further experiments showed that a protein previously identified as the microtubule-associated protein MAP-1C was in fact a motor protein that moved along microtubules in the minus end direction. Subsequent analysis demonstrated that MAP-1C is related to the dynein isolated from cilia (axonemal dynein), so MAP-1C is now referred to as cytoplasmic dynein. Kinesin is a molecule of approximately 380 kd, consisting of two heavy chains (120 kd each) and two light chains (64 kd each) (see Figure 11.45). The heavy chains have long -helical regions that wind around each other in a coiled-coil structure. The amino-terminal globular head domains of the heavy chains are the motor domains of the molecule: They bind to both microtubules and ATP, the hydrolysis of which provides the energy required for movement. Although the motor domain of kinesin (approximately 340 amino acids) is much smaller than that of myosin (about 850 amino acids), X-ray crystallography indicates that the kinesin and myosin motor domains are structurally similar, suggesting that kinesin and myosin evolved from a common ancestor. The tail portion of the kinesin molecule consists of the light chains in association with the carboxy-terminal domains of the heavy chains. This portion of kinesin is responsible for binding to other cell components (such as membrane vesicles and organelles) that are transported along microtubules by the action of kinesin motors. Dynein is an extremely large molecule (up to 2000 kd), which consists of two or three heavy chains (each about 500 kd) complexed with a variable number of light and intermediate polypeptides, which range from 14 to 120 kd (see Figure 11.45). As in kinesin, the heavy chains form globular ATP-binding motor domains that are responsible for movement along microtubules. The basal portion of the molecule, including the light and intermediate chains, is thought to bind to other subcellular structures, such as organelles and vesicles. Like the myosins, both kinesin and dynein define families of related motor proteins. Following the initial isolation of kinesin in 1985, a variety of kinesin-related proteins have been identified. Eighteen different kinesins are encoded in the genome of C. elegans, and it is thought that there may be as many as 100 different members of the kinesin family in humans. Some members of the kinesin family, like kinesin itself, move along microtubules in the plus end direction (see Figure 11.45). Other members of the kinesin family, however, move in the opposite direction, toward the minus end. Different members of the kinesin family vary in the sequences of their carboxy-terminal tails and are responsible for the movements of different types of "cargo," including vesicles, organelles, and chromosomes, along microtubules. There are also several types of axonemal dynein, as well as multiple cytoplasmic dyneins. All members of the dynein family move toward the minus ends of microtubules, but different cytoplasmic dyneins may transport different cargoes. Organelle Transport and Intracellular Organization One of the major roles of microtubules is to transport membrane vesicles and organelles through the cytoplasm of eukaryotic cells. As already discussed, such cytoplasmic organelle transport is particularly evident in nerve cell axons, which may extend more than a meter in length. Ribosomes are present only in the cell body and dendrites, so proteins, membrane vesicles, and organelles (e.g., mitochondria) must be transported from the cell body to the axon. Via video-enhanced microscopy, the transport of membrane vesicles and organelles in both directions can be visualized along axon microtubules, where kinesin and dynein carry their cargoes to and from the tips of the axons, respectively. For example, secretory vesicles containing neurotransmitters are carried from the Golgi apparatus to the terminal branches of the axon by kinesin. In the reverse direction, cytoplasmic dynein transports endocytic vesicles from the axon back to the cell body. Microtubules similarly transport membrane vesicles and organelles in other types of cells. Because microtubules are usually oriented with their minus end anchored in the centrosome and their plus end extending toward the cell periphery, different members of the kinesin and dynein families are thought to transport vesicles and organelles in opposite directions through the cytoplasm (Figure 11.46). Figure 11.46. Transport of vesicles along microtubules Kinesin and other plus end-directed members of the kinesin family transport vesicles and organelles in the direction of microtubule plus ends, which extend toward the cell periphery. In contrast, dynein and minus end-directed members of the kinesin family carry their cargo in the direction of microtubule minus ends, which are anchored in the center of the cell. Conventional kinesin and other plus end-directed members of the kinesin family carry their cargo toward the cell periphery, whereas cytoplasmic dyneins and minus end-directed members of the kinesin family transport materials toward the center of the cell. In addition to transporting membrane vesicles in the endocytic and secretory pathways, microtubules and associated motor proteins position membrane-enclosed organelles (such as the endoplasmic reticulum, Golgi apparatus, lysosomes, and mitochondria) within the cell. For example, the endoplasmic reticulum extends to the periphery of the cell in association with microtubules. Drugs that depolymerize microtubules cause the endoplasmic reticulum to retract toward the cell center, indicating that association with microtubules is required to maintain the endoplasmic reticulum in its extended state. This positioning of the endoplasmic reticulum appears to involve the action of kinesin (or possibly multiple members of the kinesin family), which pulls the endoplasmic reticulum along microtubules in the plus end direction, toward the cell periphery. Similarly, kinesin appears to play a key role in the positioning of lysosomes away from the center of the cell, and three different members of the kinesin family have been implicated in the movements of mitochondria. Conversely, cytoplasmic dynein is thought to play a role in positioning the Golgi apparatus. The Golgi apparatus is located in the center of the cell, near the centrosome. If microtubules are disrupted, either by a drug or when the cell enters mitosis, the Golgi breaks up into small vesicles that disperse throughout the cytoplasm. When the microtubules re-form, the Golgi apparatus also reassembles, with the Golgi vesicles apparently being transported to the center of the cell (toward the minus end of microtubules) by cytoplasmic dynein. Movement along microtubules is thus responsible not only for vesicle transport, but also for establishing the positions of membrane-enclosed organelles within the cytoplasm of eukaryotic cells. Separation of Mitotic Chromosomes As discussed earlier in this chapter, microtubules reorganize at the beginning of mitosis to form the mitotic spindle, which plays a central role in cell division by distributing the duplicated chromosomes to daughter nuclei. This critical distribution of the genetic material takes place during anaphase of mitosis, when sister chromatids separate and move to opposite poles of the spindle. Chromosome movement proceeds by two distinct mechanisms, referred to as anaphase A and anaphase B, which involve different types of spindle microtubules and associated motor proteins. Anaphase A consists of the movement of chromosomes toward the spindle poles along the kinetochore microtubules, which shorten as chromosome movement proceeds (Figure 11.48). Figure 11.48. Anaphase A chromosome movement Chromosomes move toward the spindle poles along the kinetochore microtubules. Chromosome movement is thought to be driven by minus enddirected motor proteins associated with the kinetochore. The action of these motor proteins is coupled to disassembly and shortening of the kinetochore microtubules. This type of chromosome movement appears to be driven principally by kinetochore-associated motor proteins that translocate chromosomes along the spindle microtubules in the minus end direction, toward the centrosomes. Cytoplasmic dynein is associated with kinetochores and may play a role in poleward chromosome movement, as may minus end-directed members of the kinesin family. The action of these kinetochore motor proteins is coupled to disassembly and shortening of the kinetochore microtubules, which may be mediated by some members of the kinesin family that act as microtubule-destabilizing enzymes. Anaphase B refers to the separation of the spindle poles themselves (Figure 11.49). Figure 11.49. Spindle pole separation in anaphase B The separation of spindle poles results from two types of movement. First, overlapping polar microtubules slide past each other to push the spindle poles apart, probably as a result of the action of plus end-directed motor proteins. Second, the spindle poles are pulled apart by the astral microtubules. The driving force could be either a minus enddirected motor anchored to a cytoplasmic structure, such as the cell cortex, or a plus end-directed motor associated with the spindle pole Spindle-pole separation is accompanied by elongation of the polar microtubules and is similar to the initial separation of duplicated centrosomes to form the spindle poles at the beginning of mitosis (see Figure 11.43). During anaphase B the overlapping polar microtubules slide against one another, pushing the spindle poles apart. This type of movement has been found to result from the action of several plus end-directed members of the kinesin family, which crosslink polar microtubules and move them toward the plus end of their overlapping microtubule away from the opposite spindle pole. In addition, the spindle poles may be pulled apart by the astral microtubules. The mechanism responsible for this type of movement has not been established, but it could result from the action of cytoplasmic dynein anchored to the cell cortex or another structure in the cytoplasm. The translocation of such an anchored dynein motor along astral microtubules in the minus end direction would have the effect of pulling the spindle poles apart, toward the periphery of the cell. Alternatively, a motor protein associated with the spindle poles could move along astral microtubules in the plus end direction, which would also pull the spindle poles toward the cell periphery. Cilia and Flagella Cilia and flagella are microtubule-based projections of the plasma membrane that are responsible for movement of a variety of eukaryotic cells. Many bacteria also have flagella, but these prokaryotic flagella are quite different from those of eukaryotes. Bacterial flagella (which are not discussed further here) are protein filaments projecting from the cell surface, rather than projections of the plasma membrane supported by microtubules. Eukaryotic cilia and flagella are very similar structures, each with a diameter of approximately 0.25 m. Many cells are covered by numerous cilia, which are about 10 m in length. Cilia beat in a coordinated back-and-forth motion, which either moves the cell through fluid or moves fluid over the surface of the cell. For example, the cilia of some protozoans (such as Paramecium) are responsible both for cell motility and for sweeping food organisms over the cell surface and into the oral cavity. In animals, an important function of cilia is to move fluid or mucus over the surface of epithelial cell sheets. A good example is provided by the ciliated cells lining the respiratory tract, which clear mucus and dust from the respiratory passages. Flagella differ from cilia in their length (they can be as long as 200 m) and in their wavelike pattern of beating. Cells usually have only one or two flagella, which are responsible for the locomotion of a variety of protozoans and of sperm. The fundamental structure of both cilia and flagella is the axoneme, which is composed of microtubules and their associated proteins (Figure 11.51). Figure 11.51. Structure of the axoneme of cilia and flagella (B) microtubule, containing only 10 or 11 protofilaments. The outer doublets are joined to each other by nexin links and to the central pair of microtubules by radial spokes. Each outer microtubule doublet is associated with inner and outer dynein arms. The microtubules are arranged in a characteristic "9 + 2" pattern in which a central pair of microtubules is surrounded by nine outer microtubule doublets. The two fused microtubules of each outer doublet are distinct: One (called the A tubule) is a complete microtubule consisting of 13 protofilaments; the other (the B tubule) is incomplete, containing only 10 or 11 protofilaments fused to the A tubule. The outer microtubule doublets are connected to the central pair by radial spokes and to each other by links of a protein called nexin. In addition, two arms of dynein are attached to each A tubule, and it is the motor activity of these axonemal dyneins that drives the beating of cilia and flagella. The minus ends of the microtubules of cilia and flagella are anchored in a basal body, which is similar in structure to a centriole and contains nine triplets of microtubules. Centrioles were discussed earlier as components of the centrosome, in which their function is uncertain. Basal bodies, however, play a clear role in organization of the axoneme microtubules. Namely, each of the outer microtubule doublets of the axoneme is formed by extension of two of the microtubules present in the triplets of the basal body. Basal bodies thus serve to initiate the growth of axonemal microtubules, as well as anchoring cilia and flagella to the surface of the cell. The movements of cilia and flagella result from the sliding of outer microtubule doublets relative to one another, powered by the motor activity of axonemal dynein (Figure 11.53). Figure 11.53. Movement of microtubules in cilia and flagella The bases of dynein arms are attached to A tubules, and the motor head groups interact with the B tubules of adjacent doublets. Movement of the dynein head groups in the minus end direction (toward the base of the cilium) then causes the A tubule of one doublet to slide toward the base of the adjacent B tubule. Because both microtubule doublets are connected by nexin links, this sliding movement forces them to bend. The dynein bases bind to the A tubules while the dynein head groups bind to the B tubules of adjacent doublets. Movement of the dynein head group in the minus end direction then causes the A tubule of one doublet to slide toward the basal end of the adjacent B tubule. Because the microtubule doublets in an axoneme are connected by nexin links, the sliding of one doublet along another causes them to bend, forming the basis of the beating movements of cilia and flagella. It is apparent, however, that the activities of dynein molecules in different regions of the axoneme must be carefully regulated to produce the coordinated beating of cilia and the wavelike oscillations of flagella a process about which little is currently understood