Nucleosides, Nucleotides & Nucleic Acids Nucleotides Nitrogenous
advertisement

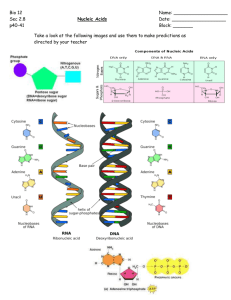
Nucleosides, Nucleotides & Nucleic Acids • Genetic (heritable) information of the cell • Also have roles as – energy transfer molecules – cofactors/coenzymes 1 Nucleotides • Building block of the nucleic acids • 3 characteristic components – Nitrogen containing base – Pentose sugar – Phosphoryl group • If no phosphoryl group, then nucleoside 2 Nitrogenous bases • Heterocyclic ring • Two parent components – Purine – Pyrimidine • Sugar link at N – N9 purines – N1 pyrimidines 3 1 4 Pentose Sugar • Pentose sugar in β-furanose form • Numbered by carbohydrate convention – Distinguished from base numbering by ‘prime’ (´) • Ring is puckered • Linked to the nitrogenous base at C1´ position – N-β-glycosidic bond • Classification based upon which sugar is present – Ribose or deoxyribose 5 Phosphate Group • Nucleotides have anywhere from 1 to 3 phosphoryl groups • Linked to sugar • Named by number of phosophates – Mono-, di-, tri- • Position of bonds identified by sugar position – 5’ – 2’3’ cyclic monophosphate 6 2 7 8 9 3 10 11 Cellular Roles • Nucleic acid constituents – Genetic information – RNA, DNA • Components of enzyme cofactors • Energy currency • Act as messengers – Link cellular responses to extracellular stimuli (I.e.hormones) 12 4 Nucleic Acids • Covalently linked nucleotides by phosphoryl group bridges • The 5’ phosphate linked to 3’ hydroxyl of next nucleotide – Phosphodiester linkages – backbone is alternating phosphate and pentose residues – Nitrogenous bases, “side-group”, face out at regular intervals • Phosphate is completely ionized • Sugar hydroxyl H-bonded to water • Nitrogenous bases are hydrophobic 13 Nomenclature • Oligonucleotide – Normally less than 50 bases • Polynucleotide – Termed nucleic acid 14 15 5 Directional molecule • Phosphodiester links have same orientation giving the linear nucleic acid specific polarity • Schematic representation via line drawing 16 DNA vs RNA • Based upon the identity of the sugar • Both subject to slow hydrolysis of the phosphodiester bond • Basic conditions will rapidly hydrolyze RNA – Hydroxyl on 2’ position makes 3’ phosphodiester susceptible to hydrolysis 17 18 6 Structure/Composition • DNA stores genetic information • 1868 first isolation of “nuclein” – Fredrich Mieschner suspected that it was responsible for inheritance • 1st direct evidence of DNA being heritable information molecule – Avery-MacLeod-McCarty experiment 1944 – Hershey-Chase experiment 1952 19 Chargaff’s Rules • Distinctive base composition noted • Proposed in 1940’s – Base composition of DNA varies by species – DNA from different tissues, but same species same – Base composition doesn’t change due to nutrition/ age/environment • Regardless of species, for all cellular DNA – A=T; C=G – A+G=C+T 20 • Non-direct evidence of DNA inheritance 21 7 Hershey-Chase • Identification of which part of viral particle infects the cell • Direct evidence of material transferred 22 Watson Crick Double Helix Structure • Knew prior information – Chargaff’s rules – Rosaline Frank/Maurice Wilkins X-ray diffraction pattern • Helices, two perodices along long axis • One 3.4 Å, 2nd 34 Å • Proposed a model that corelated all the information 23 Model proposed in 1953 • 2 helical chains wound around same axis – Right-handed double helix • Hydrophilic backbone alternating deoxyribose and phosphate groups on exterior – C2’ endo conformation of sugar • Hydrophobic and nearly planer nucleotide base close together – perpendicular to long axis • Pairing yields major and minor grooves 24 8 • Parallel or antiparallel strands? – Antiparallel yields two complementary structures • when A on one strand, T on opposite – The complement • Double helix held together by: – hydrogen bonding between bases – base stacking interactions of the ring structures • Hydrophobic interactions of the planer rings 25 26 9 28 29 3 forms of DNA • DNA is a relatively flexible molecule • Flexibility due to – Possible conformations of the deoxyribose ring – Rotation about the contiguous bonds that make up the phosphodiester backbone – Free rotation about the C1´-N glycosyl bond • 3 structural forms predominate – A,B and Z 30 10 B-form DNA • Watson-Crick proposed model • Most stable under physiological conditions • Standard reference molecule • Physical parameters – 10.5 bases per turn – 3.4 Å rise per base – 34 Å per turn – Right-handed helix 31 A-form DNA • Favored in solutions devoid of water • Physical parameters – 11 bases/turn – Base plane tilted • Deepened major groove • Shallower minor groove • Uncertain if found physiologically 32 Z-form DNA • Radical departure from prior structures • Physical parameters – left-handed helix – 12 bp/turn – 2.7 Å rise per base – Elongated (zig-zag) appearance – Purine residues flip to the syn conformation • Found, role is uncertain 33 11 34 Palindromic sequences • Word or phrase that reads the same forwards or backwards – “Level, madam, level!" 35 RNA structure • Complex structures exist for RNA molecules • RNA molecule has more functions within the cell – mRNA - messenger – rRNA - ribosomal – tRNA - transfer 36 12 mRNA • Messenger RNA carries genetic message from the nucleus to the cytoplasm • mRNA from different genes vary in length, mRNA from a single gene has defined size – Monocistronic vs polycistronic 37 • Stem-loop structures – Both double and single stranded areas 38 rRNA • Ribosomal RNA • Jacob-Monod proposal – ribosomes were not manufactured anew each time a protein was made – the ribosomes did not contain the template necessary for the manufacture – Were structures that when supplied with the necessary building blocks and instruction (mRNA) • Conserved within organisms – Most conserved gene – 16S/18S RNA allows taxonomic identification 39 13 40 tRNA • transfer RNA • Adaptor molecule allowing interaction between codon (nucleotide sequence) and amino acid • Has specific 3-dimensional structure – Acceptor stem – Anticodon stem-loop – TΨC loop – D (dihydrouridine) loop 41 42 14 RNA and DNA can be denatured • Denaturation - removal of Hbonding – Temperature, pH – No covalent bonds broken • Renaturation called annealing • One step – Rapid if molecules still associated at some point – “Zipping-up” of molecule • Two step – Slower, molecules not associated – First must associate, then have regions of complementarity before the zip can occur 43 Tm • Temperatures are content specific • G-C higher melting temp than A-T • Greater number of Hbonds 44 Transformation • Spontaneous loss of groups – Deamination • exocyclic amino group – Depurination • Loss of nitrogenous bases • Hydrolysis of N-b-glycosyl bond higher for purines than pyrimidines • Pyrimidine dimers induced by UV irradiation – Formation of a cyclobutyl ring between two adjacent thymines • Methylation – Addition of a methyl (-CH3) group – Adenine and cytodine more often methylated – S-adenosylmethionine methyl group donor • Mutation – If in DNA, daughter chains will have permanent change45to the sequence 15 46 47 Sanger dideoxy sequencing • Requires – Template – Primer – All four dNTPs + ddNTPs radiolabeled – Enzyme (polymerase) • Label incorporated is complementary to base present on template 48 16 49 Energy Currency • Phosphoanhydride bond between phosphoryl groups in nucleotidetriphosphates is a high energy bond – Have α, β, γ phosphoryl groups – The α-β, β-γ are phosphoanhydride – Sugar to α is a ester • Most common carrier of energy is the adenine base 50 Enzyme cofactors • Have nucleotide as part of the structure • Nucleotide doesn’t participate in the function, but acts as a handle, allowing binding energy to participate in enzyme-substrate interaction 51 17 Regulatory molecules/messengers • Serve to modulate the activity of enzymes/ pathways • Second messengers as they are produced in the cell in response to message from outside of the cell • Commonly cyclic AMP (cAMP, 3’5’cAMP) and ppGpp 52 18