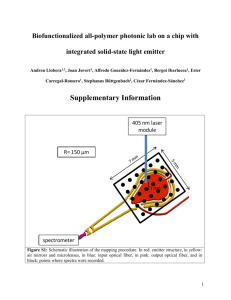
determination of the concentration of an
advertisement

DETERMINATION OF THE CONCENTRATION OF AN UNKNOWN PROTEIN SOLUTION LAB PRO 1 INTRODUCTION Cellular proteins are essential to the functioning of a healthy cell. They are responsible for processes as diverse as the replication of DNA or the detection of an invading virus. One way scientists study the function of an individual protein is to extract it from the cell and isolate that single protein from all the other cellular proteins. This process, known as protein purification, involves many steps. At each step, there is the chance of technical or human error, both of which can affect the overall yield (amount) of protein recovery. With this type of variability, it is essential to determine the final concentration of each Figure 1. Amino Acids Bound by Dye BASIC AMINO ACIDS H H O H N+ H C C H O CH2 N + C CH3 - H CH2 H O CH2 N - + O H C C O- CH2 NH CH2 + CH2 H3N+ H O NH2 CH2 H2N CH2 Lysine (Lys or K) C NH NH Arginine (Arg or R) Histidine (His or H) AROMATIC AMINO ACIDS H H H N + H C CH2 H C O- N + H H O O C CH2 C H O - N + H O C CH2 C ONH OH Phenylalanine (Phe or F) Westminster College SIM Tyrosine (Tyr or Y) Tryptophan (Trp or W) PRO1-1 Determination of the Concentration of an Unknown Protein Solution purification procedure. Determination of the concentration is usually done by comparing the protein solution to a standard curve. Once the concentration is determined, this information can be correlated with an enzyme assay to give an overall picture of the activity and purity of a particular protein purification procedure. Proteins, or polypeptides, are made of amino acids joined by peptide bonds. The protein concentration assay used in this lab is based upon an acidic dye that will bind to basic and aromatic amino acid residues. The amino acids that would be bound by the dye are arginine, lysine, histidine, tyrosine, tryptophan and phenylalanine (Fig. 1). Alone, the acidic dye has a maximum absorbance at a wavelength of 465 nanometers (nm). When bound to protein, the maximum absorbance of the dye shifts to 595 nm. The dye also has a differential color change in response to various concentrations of protein. This means that the more protein there is in a solution, the darker the color of the bound dye. Both the absorbance shift and the color change are detectable using a visible light spectrophotometer. A standard curve is created by testing a protein of known concentration with the dyebinding assay to be used. In Figure 2a, the protein BSA (bovine serum albumin) is tested between the concentrations of 2 micrograms per milliliter (µg/mL) and 28 µg/mL. Within this test set, only the concentrations between 4 µg/mL and 20 µg/mL (see →’s) are considered to be in a linear range. The linear range is defined as the part of the curve where the protein concentration and the absorbance reading increase in a manner proportional to one another. Outside of this range, it is not possible for the absorbance reading to accurately reflect the concentration of an unknown solution. For example, there is very little difference in the absorbance readings of 20, 24 and 28 µg/mL, indicating this assay is unable to accurately determine protein concentrations higher than 20 µg/mL. Once the linear range is determined, these data points are used to create a linear plot. A graphing calculator or spreadsheet application (like Microsoft Excel) can determine the best-fit straight line using the least-squares method (see equation, Fig. 2b). The straight line equation produced is based on the formula: y = mx + b where y is the absorbance; x is the protein concentration in µg/mL; m is the straight line slope; and b is the straight line intercept (the value of y when x = 0). Using this equation, it is possible to determine the concentration of an unknown protein solution based on the Westminster College SIM PRO1-2 Determination of the Concentration of an Unknown Protein Solution absorbance reading (solve equation for x). The R2-value is an indicator of the precision of the standard curve, or the deviation of the concentration standards from the best-fit straight line. To achieve an acceptable level of precision, samples for the standard curve are usually performed in duplicate or triplicate. In general, an R2-value should be above 0.95 for use in determining the concentration of an unknown sample. Figure 2a. De te rmining Line ar Range 0.45 Absorbance at 595 nm 0.4 0.35 0.3 0.25 0.2 0.15 0.1 0.05 0 0 2 4 8 12 16 20 24 28 Protein concentration (ug/mL) Figure 2b. Protein Concentration Standard Curve 0.9 0.8 Absorbance at 595 nm 0.7 0.6 0.5 y = 0.0413x – 0.0061 0.4 R2 = 0.9986 0.3 0.2 0.1 0 -0.1 0 5 10 15 20 25 Protein concentration (ug/mL) Westminster College SIM PRO1-3 Determination of the Concentration of an Unknown Protein Solution PURPOSE This purpose of this laboratory exercise is to determine the concentration of two unknown protein solutions. This procedure involves a dye-binding protein assay with known amounts of the protein BSA (bovine serum albumin) and the use of a visible light spectrophotometer to measure the assay results and to create a standard curve. Utilizing the standard curve, students will calculate the concentration of two unknown protein solutions. EQUIPMENT/MATERIALS Spectronic 20 Genesys disposable plastic cuvettes 2-20 µL pipettor 200-1000 µL pipettor 1.5 mL Eppendorf tubes 2 unknown protein solutions BioRad Protein Assay Dye Reagent wash bottle with deionized water Kimwipes 2-20 µL pipet tips 200-1000 µL pipet ti Eppendorf racks 20 mg/mL BSA protein solution goggles and gloves SAFETY Gloves and goggles should be worn during this lab, especially when handling the dye reagent. This dye contains phosphoric acid and methanol, both of which will irritate the skin and eyes. PROCEDURE Preparing the Spectronic 20 Genesys 1. Turn on the spectrophotometer. The spectrophotometer should warm up for at least 10 minutes. Use the same instrument for the entire experiment. 2. Set the spectrophotometer to 595 nm using the ▲nm and ▼nm buttons. Creating a Standard Curve 1. A 1 mg/mL BSA standard solution is provided. This protein will be used to create a standard curve that spans a range of 2-20 µg/mL. Obtain fourteen 1.5 mL Eppendorf tubes. Label two “0”, two “2”, two “4”, etc. to match the number of µL BSA added, as indicated in Table 1. The numbers in the BSA column also reflect the µg/mL in each sample. Every sample must be done in duplicate to ensure an accurate standard curve. Westminster College SIM PRO1-4 Determination of the Concentration of an Unknown Protein Solution Table 1 Preparation of BSA standard curve samples µL BSA standard µL distilled water µL dye reagent 0 800 200 2 798 200 4 796 200 8 792 200 12 788 200 16 784 200 20 780 200 2. Using the values in Table 1, mix the appropriate amount of water and BSA standard using the micropipetors and pipet tips provided. Each BSA/water sample should have a final volume of 800 µL. Close the tube caps tightly and shake the samples vigorously to thoroughly mix the samples. Set the standard curve samples aside. DO NOT ADD THE DYE REAGENT AT THIS TIME! 3. Obtain 2 unknown protein samples from the instructor. These will be provided in duplicate, and are already diluted to a final volume of 800 µL. Record the number of the sample on the data sheet provided. 4. Once the BSA standards and unknown samples are prepared, add the 200 µL dye reagent to every tube, both the standard curve and the unknown protein samples. Cap the tubes securely and mix well by shaking. Let the samples incubate for at least 5 minutes. While this incubation is underway, obtain seven clean cuvettes and label them 0, 2, 4, 8, 12, 16, and 20, one for each of the BSA standards. 5. After 5 minutes, a color change should have occurred, resulting in a range of blue coloration depending on protein concentration. If no color difference is apparent in the standard curve samples, see the instructor. 6. The “0 µg” sample is considered the blank for this experiment. Using a disposable pipet bulb, transfer the entire sample into the cuvette labeled “0”. Place the blank cuvette into the sample compartment of the spectrophotometer with the triangle on the cuvette facing the front of the instrument. Note: Before inserting a cuvette into the spectrophotometer, wipe it clean and dry with a kimwipe, and make sure that the solution is free of bubbles. Do not touch the clear sides of the cuvette. 7. Press the “0 ABS/100%T” button on the spectrophotometer. By doing this, any background absorbance from the dye and buffer in your samples is subtracted away. (This is equivalent to buying strawberries by the pound. When you go to the field to Westminster College SIM PRO1-5 Determination of the Concentration of an Unknown Protein Solution pick the berries, you weigh the containers first. Then you subtract that weight from the filled container, so that you pay for only the berries.) 8. Record a 0 in the data table in the row for 0 BSA standard under the Absorbance A column. Remove the “0” cuvette from the instrument. 9. Transfer the “2” sample to the cuvette labeled 2. Place the cuvette containing the “2” sample into the spectrophotometer. Make sure that the triangle on the cuvette is facing the front of the instrument. Do not press 0 ABS 100%T. Record the absorbance reading in the data table row for BSA standard 2 in the Absorbance A column. 10. Continue to take readings for samples 4 – 20, recording the absorbance in the Absorbance A column. 11. Repeat Steps 6-10 for the duplicate samples of the BSA standard. You may pour the first set of standards into the sink and reuse the labeled cuvettes for the duplicate standards. Record these absorbance readings in the column labeled Absorbance B on the Data Sheet. 12. Once you have recorded the data for the standard curve, repeat Steps 6 – 10 for the protein samples of unknown concentration. Again, these samples should be provided to you in duplicate. Record the absorbance readings in the table provided on the Data Sheet. 13. Plot the results of the standard curve using the graph paper provided. The x-axis is BSA concentration (µg/mL) and the y-axis is the average absorbance readings at 595 nm. All these samples are within the linear range of this particular assay, so if the precision for the standard curve is good, then connecting the data points should approximate a straight line. (The standard curve may also be graphed on a graphing calculator or a spreadsheet program, if these are available.) 14. Determine the linear equation using a graphing calculator or a spreadsheet application. If neither of these is available, see your instructor. NOTE: It is possible obtain a linear best-fit without a calculator or spreadsheet. To create a best-fit line, make a scatter plot with the data points obtained from the standard curve. Instead of connecting the points, estimate the best place to draw a line between all the data points. The intersection of the absorbance reading and the hand-drawn line will indicate the approximate concentration of the unknown protein solution (see Fig. 3). Westminster College SIM PRO1-6 Determination of the Concentration of an Unknown Protein Solution Figure 3. Hand-Drawn Best-Fit Line 1.0 * 0.8 Absorbance * * 0.6 0.4 0.2 * = data points = best-fit line * * * * 2 4 6 8 10 12 14 BSA Std (µg/mL) In the above example, the absorbance reading for the unknown sample is 0.55. The point where this absorbance intersects the best-fit line indicates what the protein concentration is for the unknown sample (~9.3 µg/mL). References: Adapted from “Bio-Rad Protein Assay” Instruction Booklet, 2004. Additional Reference: James S. Fritz and George H. Schenk. Quantitative Analytical Chemistry. “Appendix 5: Least-Squares Method for Linear Plots”. Allyn and Bacon, Inc. 1979. 4th Edition. p. 638. Westminster College SIM PRO1-7 Determination of the Concentration of an Unknown Protein Solution DATA SHEET Name ________________________ Name ________________________ Period _______ Class ___________ Date ___________ DETERMINATION OF THE CONCENTRATION OF AN UNKNOWN PROTEIN SOLUTION DATA TABLES DATA TABLE 1 STANDARD CURVE ABSORBANCE DATA BSA standard: Absorbance A Absorbance B Average Absorbance 0 2 4 8 12 16 20 DATA TABLE 2 UNKNOWN PROTEIN CONCENTRATION ABSORBANCE DATA Unknown Protein #: Absorbance A Absorbance B Average Absorbance Linear Equation (y = mx + b): ______________________ R2 Value: _______ Westminster College SIM PRO1-8 Determination of the Concentration of an Unknown Protein Solution Use the linear equation to calculate the protein concentration of both unknown samples. Unknown Protein # ___: Concentration: ________µg/mL Unknown Protein # ___: Concentration: ________µg/mL Westminster College SIM PRO1-9 Determination of the Concentration of an Unknown Protein Solution QUESTIONS Name ________________________ Period _______ Class ___________ Date ___________ 1. a. How precise was your BSA standard curve (what was the R2-value)? b. What are possible sources of error (technical or human) that affect the R2-value and how could they be corrected? 2. Why is it necessary to determine the concentration of every protein purification? 3. What is the linear range on a protein concentration curve? 4. Did either of your unknown samples have an absorbance reading outside the linear range of your standard curve? If so, can you accurately determine the concentration of the sample? Explain why or why not. 5. Why is it necessary to have a “0” sample, with no protein, as part of the standard curve? Westminster College SIM PRO1-10 Determination of the Concentration of an Unknown Protein Solution Westminster College SIM PRO1-11