Chromosome Mapping in Sordaria
advertisement

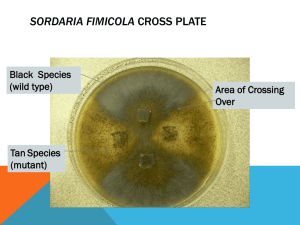
AP Biology Lab: Chromosome mapping by frequency of crossing over (tetrad analysis) in Sordaria fimicola Sordaria fimicola is an ascomycete fungus (as is Neurospora crassa). Ordinarily these organisms are haploid, and grow to form filaments (hyphae) that form a tangled mat called a mycelium. However, these haploid organisms can engage in sexual reproduction, resulting in a transient diploid condition. That happens when hyphae from different organisms fuse, which introduces haploid nuclei from each into the same cytoplasm. These nuclei fuse to produce a diploid nucleus, which then immediately undergoes meiosis to produce 4 haploid nuclei. Each haploid nucleus undergoes one mitotic division, resulting in eight nuclei that become encased in individual ascospores (each of which can germinate into a new haploid organism). These meiotic and mitotic divisions occur in a cell that gradually elongates to accommodate the increasing number of nuclei, such that all nuclei are lined up in single file. This situation means that the order of the nuclei in each cell (ascus) reflects the lateral arrangement of homologous pairs formed during meiosis. In Sordaria, there are several asci contained inside specialized reproductive structures called perithecia. Now, as I’m sure you remember fondly, alleles that are not involved in crossover events segregate during meiosis I, whereas alleles that are involved in crossovers segregate during meiosis II. Think about it. So, if you were to observe the genotypes of the resulting ascospores (or the organisms derived from them) you could determine if segregation occurred during the first meiotic division (no crossover) or the second meiotic division (crossover). Segregation during meiosis I should produce a 4:4 arrangement of ascospore genotypes. Segregation during meiosis II should produce a 2:2:2:2 or a 2:4:2 arrangement. Yeah, so? I hear you saying. Well, the so is that you can map chromosomal loci relative to the centromere by determining the frequency of second division segregations. The farther away a locus is from its centromere, the more frequently crossing over can occur, and hence the more frequent will be second division segregations. So, you can easily do mapping in this way. Isolating ascospores and growing them up to observe phenotypes would be a laborious process. Fortunately for you, however, there are a few genes in Sordaria that affect ascospore pigmentation directly. So, for these genes you can observe segregation patterns directly by examining the different-colored ascospore arrangement in each ascus. Procedure: 1. Each group should obtain a crossing agar plate. On the bottom, divide it into quarters with a wax pencil. Mark alternate quarters +,M,+,M (see below). Also put on your group identifier. + M M + G. Wurst 2008 from http://webpub.allegheny.edu/employee/g/gwurst/Bio325/ 2. Obtain the wild type and a mutant (gray or tan) stock culture plates. Using sterile technique, remove a ~ 5 mm circular block of mycelium of the mutant culture and place it in the center of one of the “M” quadrants. Repeat this step until all four quadrants have the appropriate block of mycelium in their centers. 3. Put the plates you’ve just made at room temperature and in the dark. Observe them after about 4 days, and then daily until the perithecia are mature. 4. What’s happening is that the hyphae from each mycelial block are growing out and toward hyphae from the other blocks. Where they meet they will fuse and form mating structures culminating in black dots, the perithecia. So you should see on your plates a rough “X” of black dots 5. Observing involves (using sterile technique, of course!) removing a few perithecia with a sterilized needle, placing them in a drop of water on a slide, and covering them with a coverslip. If tapping on the coverslip with a pencil eraser or your thumb easily breaks the perithecia open and releases lots of asci with mature (observable) ascospores, the cross is ready to score. If tapping doesn’t break open the perithecia, or if there are only a few asci released which contain underdeveloped ascospores, allow the crosses to mature for another day. 6. When the perithecia are mature, continue with the experiment. If it’s inconvenient to continue when the perithecia are mature, refrigerate the cross plate until you are ready to continue the experiment. 7. When the perithecia are mature, prepare them as above. Count the number of asci with the 4:4 first division segregation pattern and the number with a 2:2:2:2 or 2:4:2 arrangement. Record these numbers. Count a total of at least 100 asci that contain both light and dark ascospores. Don’t count asci with only light or dark ascospores. Make a note of any apparently anomalous results, i.e. a 5:3, 3:1:1:3 or 6:2 pattern. Anybody who finds one of those should photograph it, save the plate and will receive a prize provided you can explain what is causing it. What would patterns like this indicate? 8. Now, calculate the map distance from the centromere to the gray (or tan) locus by dividing the number of second division segregation asci (tetratypes) by the total number of asci scored (tetratypes + ditypes). Multiply by 100 to obtain a percentage (i.e. map units) then divide by 2 to get the map distance. Why do you divide by 2? G. Wurst 2008 from http://webpub.allegheny.edu/employee/g/gwurst/Bio325/