Restriction Digestion and Analysis of Lambda DNA Kit (BioRad)
advertisement

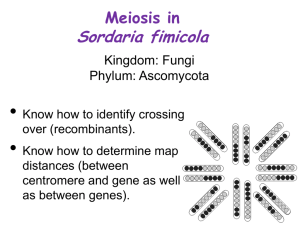
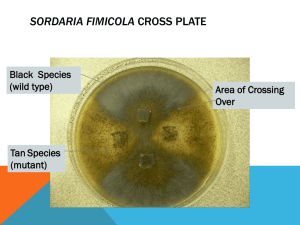
Sordaria Genetics BioKit (Carolina) Introduction With the Sordaria kit, you will observe the results of crossing over between wild-type Sordaria fimicola with mutant tan and between wild-type and mutant gray. The results are then used to calculate the gene-to-centromere distance in map units. Background Sordaria fimicola is a common ascomycete that grows on the dung of herbivores. Although Sordaria is homothallic, different types will form hybrids by fusion of hyphae (hyphal anastomosis). This results in hyphae containing nuclei of both strains as the vegetative hyphae are not divided by cross walls into separate cells. A crozier (shepherd’s crook) develops at the end of an ascogenous hyphae, and the two nuclei within the crozier simultaneously divide to produce four nuclei. Two of these nuclei (one from each parent nucleus) are cut off from the rest of the hyphae by cross wall formation and fuse to form a diploid cell. This diploid nucleus undergoes meiosis to form four haploid nuclei). Each of these nuclei then undergo mitosis, producing a total number of eight nuclei, which become ascospores. The plane of each division is parallel to the length of the ascus, so the final arrangement of ascospores reflects the order of segregation in meiosis. This allows the use of different spore color alleles to illustrate crossingover. As the rate of crossing-over increases with the distance between any two points on a chromosome, it is possible to determine the relative distance from a gene locus to its centromere. Such map distances originally were determined for organisms such as the fruit fly and were based upon random analysis of offspring rather than on percentages of tetrads. In Sordaria, each MII ascus represents a tetrad of chromatids in which there has been an exchange involving only two of the four chromatids. Therefore, the percentage of MII asci must be divided by two to give a map distance comparable to that determined for fruit flies and most other organisms. For example, if 30% of the asci were MII, the gene-to-centromere distance would be 15 map units. Procedure When the cross plate cultures are mature, use a toothpick to remove several perithecia (observed as dark spots near edge of plates). Make a wet mount of the perithecia and gently press the coverslip with your thumb or an eraser until the perithecia are crushed. This will release clusters of asci. Using low power of a microscope, search for hybrid asci (light and dark spores). If you are unable to find any, you may need to sample a different area of the cross plate. (regions most likely to yield hybrid asci will be discussed in class). After locating hybrid asci, use high dry magnification to count the number of MI and MII asci. Count a total of at least 300 asci for the wild-type and tan cross and record the results in the table provided. Then calculate the percent of MII and the gene-to-centromere distance in map units. Wild-Type # of MI asci # of MII asci Total asci % MII (# of Gene-toand tan (4:4) (2:4:2 or MII/Total) Centromere cross 2:2:2:2) Distance (picture #) (%MII/2) Total numbers Student questions (make sure to include a TYPED copy of the above table) 1. Why is Sordaria a useful organism to study meiosis? 2. Some perithecia have asci that contain either all 8 black or all 8 gray ascospores. How could these arrangements be explained? 3. Why does the final arrangement of ascospores reflect the order of segregation during meiosis? 4. Does the rate of crossing over increase or decrease as the distance between any two points on a chromosome increase? Explain. 5. What is a map unit? 6. Another mutant causes gray ascospores instead of the wild type dark (or tan mutant observed here). The map unit distance between gray and wild type is 18 map units. Based in your calculations, what does this reveal about the tan and gray alleles (alleles of the same gene, or different genes)?