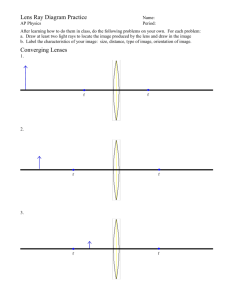
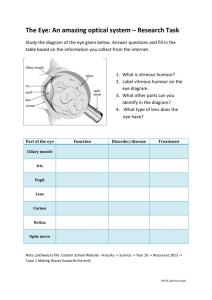
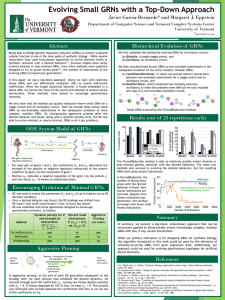
ARVO 2015 Annual Meeting Abstracts
271 Integrated genomic networks in eye development and
pathogenesis - Minisymposium
Monday, May 04, 2015 3:45 PM–5:30 PM
605/607 Minisymposium
Program #/Board # Range: 2082–2088
Organizing Section: Lens
Contributing Section(s): Biochemistry/Molecular Biology, Cornea,
Retinal Cell Biology, Retina
Program Number: 2082
Presentation Time: 3:45 PM–4:00 PM
MicroRNA networks in eye development
Ruth Ashery-Padan. Human Molecular Genetics, Tel Aviv University,
Tel Aviv, Israel.
Presentation Description: Normal vision depends on the retinal
pigmented epithelium (RPE), a monolayer of tightly connected
polarized pigmented epithelia that is physically and functionally
associated with the photoreceptor cells. Herein we investigated the
roles of microRNA (miRNA), small regulatory non-coding RNAs,
in the developing RPE. To this end we inactivated Dicer1, a key
mediator of miRNA biosynthesis, using the DctCre transgene, in
which Cre is expressed in the developing RPE after the specification.
Ablation of Dicer1 in the RPE resulted in a reduction in miRNA
typical of the RPE. The phenotype of the mutant mice was
microphthalmia, reduction in the pigmentation of the RPE, without
altered specification. Moreover the mutant mice have abnormal
development of the photoreceptors. This study reveals specific
miRNAs that are essential for maintaining the differentiation state,
adhesive properties and physiology of the RPE cells.
Commercial Relationships: Ruth Ashery-Padan, None
Support: Bright Focus Foundation
Program Number: 2083
Presentation Time: 4:00 PM–4:15 PM
Regulation of Corneal Gene Expression by Kruppel-Like Factors
KLF4 and KLF5
Shivalingappa K. Swamynathan. 1Ophthalmology, University of
Pittsburgh School of Medicine, Pittsburgh, PA; 2Cell Biology,
University of Pittsburgh School of Medicine, Pittsburgh, PA.
Presentation Description: Proper maturation and maintenance
of the stratified squamous epithelium of the cornea is essential for
normal vision. Yet, we do not completely understand the regulatory
mechanisms that controll proliferation and differentiation of the
corneal epithelial cells. Kruppel-like factors Klf4 and Klf5 are among
the top-most expressed transcription factors in the mouse cornea.
They are members of the zinc-finger family of transcription factors,
and share an identical carboxy-terminal DNA-binding domain,
while their amino-terminal regulatory domains are variable. They
both are widely expressed throughout the body, and are involved in
regulation of several critical cellular functions such as maintenance
of stem cells, formation of epithelial barrier, cell proliferation and
differentiation. Considering that the mouse Klf4- and Klf5-germline
knockouts are non-viable, we resorted to conditional disruption of
these genes in the ocular surface using Cre-Lox approach. Over
the last decade, our laboratory has described several non-redundant
functions of Klf4 and Klf5 in both maturation and maintenance of
the cornea, conjunctiva and the eyelids. We have demonstrated that
the Klf4- and Klf5-target genes are largely distinct despite their
identical DNA-binding domain, consistent with their non-redundant
roles in the mouse cornea. Considering that the expression of Klf4,
a suppressor of cell division, is more prominent in the superficial
stratified squamous epithelial cell layers of the cornea, while that
of Klf5 which promotes cell division is more prominent in the
basal cell layers, we propose that the ratio of their relative activities
determines the decision between corneal epithelial cell proliferation
and differentiation.
Commercial Relationships: Shivalingappa K. Swamynathan,
University of Pittsburgh (P)
Support: NIH Grant R01EY022898; NIH Grant P30 EY08098;
unrestricted grants from Research to Prevent Blindness and the Eye
and Ear Foundation of Pittsburgh; startup funds from the Department
of Ophthalmology, University of Pittsburgh
Program Number: 2084
Presentation Time: 4:15 PM–4:30 PM
Elucidating the cis-regulatory architecture of the retina by
epigenomic profiling
Joseph C. Corbo. Pathology and Immunology, Washington
University School of Medicine, St. Louis, MO.
Presentation Description: In this talk, I present our results utilizing
a newly developed epigenomic mapping technology, ATAC-seq,
to comprehensively identify the cis-regulatory elements of three
cell types in the outer retina: rods, cones, and bipolar cells. ATACseq utilizes a transposase-based approach to tag regions of open
chromatin and can be applied to very small numbers of cells purified
by fluorescence-activated cell sorting. This approach permits us to
probe the complex interrelations between chromatin accessibility,
nucleosome positioning, and transcription factor binding binding on
a genome-wide in rare retinal cell types. These studies are part of
a larger effort to generate comprehensive, cis-regulatory maps for
all of the major cell classes in the retina. These maps will serve as a
blueprint for understanding the transcriptional networks of the retina
and the effects of non-coding variants on disease. In addition, they
will set the stage for future work that will utilize epigenomic profiling
to characterize changes that occur during the process of retinal
development and in the course of degeneration.
Commercial Relationships: Joseph C. Corbo, None
Support: NIH grant EY01882605 and HG00679002
Program Number: 2085
Presentation Time: 4:30 PM–4:45 PM
Global analysis of transcription factor action and chromatin
conformation in developing retina
Seth Blackshaw. Johns Hopkins School of Medicine, Baltimore, MD.
Presentation Description: Retinal cell types are generated
during a series of discrete temporal intervals during the course of
neurogenesis. The molecular mechanism by which retinal progenitor
cells (RPCs) successively acquire and lose competence to generate
specific cell types is largely unknown, although it is thought to be
regulated by RPC-expressed transcription factors. Lhx2, a LIM
homeodomain factor expressed in RPCs during the full course of
neurogenesis, acts in early RPCs to promote retinal identity while
repressing alternative diencephalic fates, and also plays a critical
role in inhibiting production of retinal ganglion cells. In late-stage
RPCs, in contrast, Lhx2 is essential for generation of Muller glia,
the last-born retinal cell type. To determine how Lhx2 is able to
carry out these different functions, we have used ChIP-Seq analysis
to identify direct targets of Lhx2 in early and late-stage RPCs, and
report our findings here. In parallel, we have used other approaches
to comprehensively profile changes in chromatin conformation in
RPCs during the course of retinogenesis to determine whether these
correlate with competence changes. These studies may also help
inform research aimed at producing cell-based therapies for blinding
disorders, and help ensure the efficient generation of specific cell
types that have been lost in disease.
Commercial Relationships: Seth Blackshaw, None
©2015, Copyright by the Association for Research in Vision and Ophthalmology, Inc., all rights reserved. Go to iovs.org to access the version of record. For permission
to reproduce any abstract, contact the ARVO Office at pubs@arvo.org.
ARVO 2015 Annual Meeting Abstracts
Support: NIH grant R21EY023448
Program Number: 2086
Presentation Time: 4:45 PM–5:00 PM
DNA methylation during lens and retina development
Jeff M. Gross. Molecular Biosciences, University of Texas at Austin,
Austin, TX.
Presentation Description: DNA methylation is thought to be
an important level of epigenetic regulation of gene expression in
a variety of cell types, including those of the developing retina.
However, how changes in DNA methylation facilitate the transition
from retinal progenitor cell to differentiated neuron in vivo are largely
unknown. To address this, we have performed genome-wide reduced
representation bisulfite sequencing (for cytosine-5 methylation,
5mC), reduced representation hydroxymethylation sequencing (for
5-hydroxymethyl-cytosine, 5hmC), and RNA-Seq assays on isolated
populations of proliferative retinoblasts and newly differentiated
neurons of the zebrafish retina. To perturb DNA methylation and
determine the effects on proliferation and differentiation, single,
double and compound mutants in the genes encoding zebrafish de
novo methyltransferases have been generated (dnmt3;4;5;6;7; and
8). Furthermore, two members of the tet (ten-eleven-translocation)
family of methylcytosine dioxygenases, which catalyze the
conversion of cytosine-5 methylation to 5-hydroxymethyl-cytosine,
an intermediate form potentially involved in demethylation, are
expressed unique regions of the developing retina (tet2 and tet3), and
mutants in these have also been generated.
Commercial Relationships: Jeff M. Gross, None
Support: Alcon Research Institute
Program Number: 2087
Presentation Time: 5:00 PM–5:15 PM
Reconstruction of Pax6-dependent GRNs that govern lens
development
Ales Cvekl. Albert Einstein College of Medicine, Bronx, NY.
Presentation Description: Purpose. Embryonic tissue development
requires intricate temporal and spatial control of gene expression
that is executed through specific gene regulatory networks (GRNs).
These GRNs are comprised of individual modules that contain signal
transduction pathways, binding of specific DNA-binding transcription
factors to the promoters and enhancers, non-coding RNAs and global
and local chromatin structure of co-regulated genes inside of the
cell nuclei. Pax6 has been shown to act at multiple stages of lens
development.
Methods. ChIP-seq experiments were conducted using antibodies
recognizing Pax6, RNA polymerase II, and specific core histone
modifications using chromatin prepared from newborn mouse lens
and E12.5 forebrain. RNA-seq was conducted using the wild type
lens fibers and epithelium from newborn mouse. RNA profiling data
from Pax6-/- tissues reported earlier were re-analyzed. Unbiased
analysis of Pax6-binding regions (“peaks”) was conducted using
The MEME Suite (Motif-based sequence analysis tools). The
Pax6-interacting proteins were identified by mass spectrometry and
validated by co-immunoprecipitations using lens cell nuclear extracts.
Results. Pax6-binding in chromatin showed approximately 2/3
overlap between lens and forebrain. The majority of DNA-binding
sequences correspond to the paired domain-mediated Pax6 binding
while the remaining sequences are displayed as AT-rich sequences
containing a conserved Pax6 homeodomain DNA-recognition motif
(5’-ATTA-3’). Three Pax6-dependent GRNs to be discussed in detail
include the crystallins, regulation of cell cycle exit in lens cells, and
regulation of lens fiber cell denucleation. Among the Pax6-interacting
proteins identified, chromatin remodeling enzymes previously shown
to bind Pax6 (Brg1 and p300) as well as novel proteins that harbor
histone methyltransferase activities, represent the most interesting
part of the Pax6’s lens interactome.
Conclusions. Collectively, our data provide novel insights into three
distinct processes that control lens formation. The current proteomic
studies suggest an intriguing possibility, that Pax6 recruits multiple
distinct chromatin remodelers such as SWI/SNF, ISWI, CBP/p300,
and histone methyltransferases in order to establish a specific patterns
of histone posttranslational modifications at key gene/loci that control
lens cell identity/cell-type memory and its terminal differentiation.
Commercial Relationships: Ales Cvekl, None
Support: NIH R01 EY012200, R21 EY017296, RPB
Program Number: 2088
Presentation Time: 5:15 PM–5:30 PM
Integrated approach to decipher regulatory networks in lens
development and cataract
Salil A. Lachke. Department of Biological Sciences, University of
Delaware, Newark, DE.
Presentation Description: Identification of ocular disease associated
genes has impacted therapeutic interventions and extended our
understanding of eye development and homeostasis. Disease gene
discovery in the eye, however, remains a formidable challenge.
Indeed, the majority of the 26 genes associated with non-syndromic
human pediatric cataract were identified over a period of 25 years.
The advent of genomics presents new opportunities to design
systems-based integrative approaches to facilitate disease gene
identification and to assemble gene regulatory networks (GRN)
underlying ocular development and homeostasis. Toward this goal,
we have developed a web-based publically available bioinformatics
resource tool termed iSyTE (integrated Systems Tool for Eye
gene discovery, http://bioinformatics.udel.edu/Research/iSyTE).
iSyTE is based on innovative processing and presentation of whole
genome expression datasets for specific eye tissues, and conversion
of the wealth of molecular functional data in the literature into
an interactive resource to derive and visualize “evidence-based”
GRNs. This integrated approach allows iSyTE to effectively predict
genes relevant to the biology of specific ocular tissues. As a proof
of principal, iSyTE has greatly expedited gene discovery in the
lens, leading to the identification of several new cataract associated
genes (Tdrd7, Pvrl3, Sep15, Mafg/k, Celf1) and has contributed to
the understanding of many other important regulatory pathways
(e.g. Sip1, CBP, p300, etc.). In addition, the potential for iSyTE to
provide significant advances in the basic understanding of ocular
developmental processes is realized from its identification of Tdrd7,
a conserved RNA binding protein (RBP), perturbation of which
disrupts lens gene expression and causes juvenile cataracts in
human and animal models (Lachke 2011 Science 331:1571). This
discovery has initiated the investigation of other classes of RBPs
that function in conserved post-transcriptional regulatory pathways
in vertebrate eye development. It is anticipated that investigation of
these new genes and understudied pathways will advance the etiology
of pediatric cataracts, and the integration of this new regulatory
information into iSyTE will expand the lens GRN, in turn increasing
its efficacy in future ocular disease gene discovery.
Commercial Relationships: Salil A. Lachke, None
Support: NIH R01 EY021505
©2015, Copyright by the Association for Research in Vision and Ophthalmology, Inc., all rights reserved. Go to iovs.org to access the version of record. For permission
to reproduce any abstract, contact the ARVO Office at pubs@arvo.org.