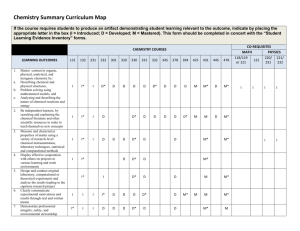
NPTEL-Module-1: Introduction to Bioorganic Chemistry Dr. S. S. Bag
advertisement

NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics NPTEL Phase – II (Syllabus Template) Course Title: Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Module II: Synthesis/Biosynthesis of Enediynes Class of Natural Products: Classifications of Natural Enediynes-Calicheamicins/ Esperamicins class of enediynes (Class I), The Dynemicins class of enediynes (Class II), and The Chromoprotein class of enediynes (Class III); Mechanism of DNA Cleavage by Each Class; Chemical Synthesis of a Few Members of Enediynes Natural Products; Biosynthesis of a Few Members of Natural Enediynes. 2.1. Classifications of Natural Enediynes 2.1.1. Introduction Enediynes are a class of bacterial natural products characterized by either nine- and tenmembered rings containing two triple bonds separated by a double bond. In the mid to late 1980s, it became clear that an emerging series of naturally occurring antitumor antibiotics such as calicheamicin, esperamicin, dynemicin, kedarcidin chromophore and C-1027 chromophore possessing the enediyne core and showed biological activity through the generation of active biradical specis via Bergman cyclization. In addition to that neocarzinostatin (NCS) chromophore which does not contain the classical conjugated enediyne system also demonstrated very similar DNA cleavage mechanism via the generation biradical species through the MyersSaito cyclization. The enediynes are in their native form biologically inactive but undergo cycloaromatization reactions after being activated by a triggering reaction and produces the active biradical species. Therefore, the enediyne group in those compounds is often called a warhead. For example, the strain imposed by the double bond in calicheamicin or by the epoxide in dynemicin imparts stability to the system. Cycloaromatization of these natural products then give rise to cytotoxic diyl radicals which are capable of inducing DNA strand scission at low concentration by abstracting –H atom from the sugar phosphate backbone of DNA. Several of the naturally occurring enediynes have entered clinical trials against cancer and in Japan neocarzinostatin is used clinically. The biological profile of the calicheamicin and esperamicins are: (a) (b) (c) (d) subpicogram potency against Gram positive bacteria, activity in the biochemical induction assay at very low concentrations, high potency against a number of animal tumor models and, induction of double-stranded DNA cleavage with minimal concurrent single strand breakage. Joint initiative of IITs and IISc – Funded by MHRD Page 1 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.1.2. Classifications of Natural Enediynes During the 1980s enediynes as a new class of natural products have been introduced with the structural elucidation of neocarzinostatin and calicheamicin. Since this time, thirteen enediynes have been structurally confirmed, which includes two probable enediynes isolated as inactive degradation products (Table 1; Figure 1). Table 1: Enediyne Natural Products Name Auromomycin Largomycin Actinoxanthin Sporamycin Neocarzinostatin C-1027 Maduropeptin Kedarcidin N1999A2 Sporolides A and B Cyanosporasides A and B Esperamicin Calicheamicin Dynemycin Namenamicin Shishijimicin Uncialamycin Producer Nine-Membered Enediynes Streptomyces macromomyceticus Streptomyces pluricolorescens Actinomyces globisporus Streptosporangium pseudovulgare Streptomyces carzinostaticus Streptomyces globisporus Actinomadura madurea Actinomycete L585-6 Streptomyces sp. AJ9493 Salinispora tropica Salinispora pacifica Ten-Membered Enediynes Actinomadura verrucosospora Micromonospora echinospora ssp. calichensis Micromonospora chersina Polysyncraton lithostrotum Didemnum proliferum Unknown Year 1968 [3] 1970 [4] 1976 [5] 1978 [6] 1985 [1] 1991 [7] 1994 [8] 1997 [9] 1998 [10] 2005 [11] 2006 [12] 1985 [13] 1987 [2] 1990 [14] 1996 [15] 2003 [16] 2005 [17] These natural antitumor antibiotics are classified under three classes: (a) The Calicheamicins and Esperamicins. (b) The Dynemicins (c) The Chromophore types; Kedarcidin chromophore, C-1027 and Neocarzinostatin. Even though these natural antitumor antibiotics possess phenomenal cytotoxicity against tumor cells they are too toxic and indiscriminant for use as drugs, hence efforts have been made to synthesize various derivatives of these compounds. A notable example is gemtuzumab ozogamicin (Mylotarg), which is a derivative of calicheamicin conjugated to a humanized antiCD33 antibody; the drug is indicated for the treatment of acute myeloid leukemia (AML). However this has been withdrawn because of its strong cytotoxicity. Joint initiative of IITs and IISc – Funded by MHRD Page 2 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Figure 1. Structures of Enediyne class of natural products. Joint initiative of IITs and IISc – Funded by MHRD Page 3 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.2. Mechanism of DNA Cleavage 2.2.1. Calicheamicins and Esperamicins The Calicheamicins (also known as the LL-E 33288 antibiotics) produced from Micromonospora echinospora spp. Calichensis, a bacterium was discovered by May. D. Lee et al., in 1987. It is the most important member of the enediyne class of natural products, and possesses potent cytotoxicity against murine tumor cells. Esperamicin A1 is also another member of the enediyne family of antibiotics exhibiting activity against marine tumor models in the 100ng/kg range. The families of Esperamicins were isolated from the bacterial Actinomadura verrucosospora and their structure elucidation was reported in 1987-89. The antitumor antibiotic drugs, calicheamicin, dynemicin, and esperamicin, all possessed bicyclo-[7,3,1]-enediyne substructure and become active p-benzyne biradical intermediates due to Bergman cyclizations. Precisely the reactive intermediate is proposed to be a 1, 4-dehydrobenzene derivative which is suggested to arise thermally from (Z)-enediyne in a cyclic version of the Bergman reaction. The mechanistic studies have revealed that at a minimum, three common features are essential to the show the potent DNA cleavage activity by these antibiotics: (a) non-destructive high-affinity binding to DNA, (b) a chemical trigging mechanism leading to a high–energy intermediate (c) rapid formation of biradical specis at physiological temperatures which is mainly responsible for DNA strand scission. Natural enediyne Calicheamicins and Esperamicins family SSSMe O SSSMe O NHCO2Me Me H HO O O OH Me O O O OH O N H O Me S HO MeO Me OMe I OMe O Calicheamicin 1I O Me O HO MeO OH HO O O O NH O OH Me O MeO MeO EtHN H O O NHCO2Me O N H OH OMe OMe NH O Me Me Esperamicin A1 Figure 2. Structures of Calicheamicin 1I and Esperamicin A1. Joint initiative of IITs and IISc – Funded by MHRD Page 4 of 89 O Me SMe NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics The esperamicins and the calicheamicins both share similar structures and their structures possess three distinct domains: (a) an oligosaccharide chain, (b) a trisulphide moiety, and (c) an enediyne core. Each of these domains has a specific function in DNA cleavage. (a) The oligosaccharide chain recognises and targets selected base pair sequences in the minor groove of DNA. Thus, the molecule binds selectively to the minor groove through hydrophobic, electrostatic interactions and hydrogen bonding of the sugar side chain with DNA. The natural enediynes are actually stable until they are bonded to DNA and then become activated. (b) After binding to minor groove, the trisulphide then serves as a molecular trigger which upon reductive activation produces thiolate. The thiolate then performs an intramolecular Michael addition onto the proximally positioned enone moiety to unlock the enediyne warhead. This leads a change in the geometry of the molecule from a trigonal bridgehead to a tetragonal centre. Thus, ―cd‖ distance between the two triple bonds is reduced. The decrease has been calculated to be from 3.35 to 3.16 Å distance which is close enough for spontaneous Bergman cyclization according to Nicolaou‘s theory. (c) Bergman cycloaromatization of the enediyne structural motif generates a p-benzyne diradical which abstracts hydrogen from DNA backbone. The reaction of the DNA backbone radicals with molecular oxygen results in double strand cleavage which ultimately lead to permanent damage of the genetic material. The enediyne systems in both the calicheamicin and esperamicin could easily be triggered to aromatize via a free-radical intermediate by cleavage at the methyl trisulfide moiety. This aromatization process is responsible for the remarkable DNA damaging effects of the calcheamicin and the esperamicins. Mechanism of DNA Cleavage by Calicheamicins Trisulfide reduction initiates the activation Intercalates into DNA Calicheamicins Responsible for DNA strand scission Scheme 1. Mechanism of DNA cleavage by Calicheamicin. Joint initiative of IITs and IISc – Funded by MHRD Page 5 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.2.2. Dynemicins Dynemicin A (DNY-A) is the first known member of the family dynemicin A 1.187. It was isolated from Micromonospora chersina M956-1109 strain and the recent member deoxydynemicin A 1.187b was obtained from Micromoonspora globosa MG331-HF6. Dynemicin contain a bicyclo[7.3.1]enediyne substructure which may be related biosynthetically to the cores of calicheamicin and esperamicin. The dynemicins has a striking hybrid structure that contains (a) the cyclic enediyne, (b) an anthraquinone chromophore. Unlike the other members of this class, it exhibits antibacterial and antitumor activity with low toxicity. As a result of their intriguing and unique structural characteristics, various strategies have been developed to provide a synthetic route towards the natural and the non-natural dynemicin and its analogues. Natural enediyne Dynemicins family OH O H HN Me CO2H O O H N PhO O OMe H R O OH R = -OH; Dynemicin A Dynemicin Model R = H; Deoxydynemicin A Figure 3. Structures of natural Dynemicin A and its model compound. Nicolaou et al., reported the synthesis of the dynemicin model compound (Figure 3) to demonstrate the mechanism of cyclization reactions of dynemicin A. In this model the critical distance (cd) was found to be 3.59 Å (popularly known as cd distance), a value that agrees with the X-ray crystallographic analysis of dynemicin A (3.54 Å). The mechanism of the cyclization reaction is as below (Scheme 2): (a) Protonation of the epoxide group initiates the formation of diol (b) Spontaneous Bergman cyclization to form benzenoid biradical. (c) Rapid trapping of the biradical by the hydrogen donor present to give the cyclized product. This cyclization is analogous to those observed for dynemicin A. The pharmacological activity of this model compound is believed to be related to dynemicin A‘s ability to cleave DNA following its intercalation into DNA with its anthraquinone which in actual fact is typical of most enediyne cyclization reactions. It is the benzenoid biradical that is actually responsible for the cleavage of the DNA molecule as illustrated in the scheme below, (Scheme 3). Joint initiative of IITs and IISc – Funded by MHRD Page 6 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Scheme 2. Mechanism of the cyclization reaction of a dynemicin model compound. Mechanism of Biological Action of Dynemicin A OH O H HN Me CO2H O OMe Dynemicin OH O OH O Me H HN HO CO2H Path A OH H OH OH HN Me CO2H Path B O OH O H HN HO OMe Me CO2H H+ OMe OMe OH OH OH OH OH OH OH OH OH H2O or Nu Proton Transfer Nucleophilic attack Me H OH OH HN HO OH O CO2H OH O OH OH OH OH OH OH Me CO2H OH OH OH OH OMe OMe Cycloaromatization CO2H DNA OMe OH DNA OH Me H OH OH HN HO Cycloaromatization H OH OH HN HO CO2H H OMe OH Me H HN HO O2 OH O Me H HN HO DNA Diradical OH O CO2H H OMe O2 DNA Double Strand Cleavage OH O H HN HO Me CO2H OH OH O OH O OH O OH DNA Double Strand Cleavage OMe OH Scheme 3. Mechanism of biological action of dynemicin A. Joint initiative of IITs and IISc – Funded by MHRD CO2H H DNA Diradical O2 Me H HN HO Page 7 of 89 OH OMe NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.2.3. The Chromoprotein Types Enediyne Class 2.2.3.1. Neocarzinostatin Chromophore (NCS) NCS is the first enediyne antibiotic that was first isolated from a culture of Streptomyces carzinostaticus var. F-41 in 1965. Its potent antibacterial and antitumor activities derived from the inhibition of DNA synthesis and DNA degradation in cells. It is composed of a very unstable chromophore and a carrier apoprotein. The neocarzinostatin core is slightly different from the basic enediyne structure. It contains the bicyclo[7.3.0]dodecenediyne and shows its biological activity through the involvement of the allene eneyne system. Meyer-Saito cyclization (MSC) is believed as the key step in the mechanism of action of the antitumor agent neocarzinostatin chromophore through which it produced a 3, 7-dehydroindene derivative as shown below (Scheme 4). The generated biradical via MSC is responsible to abstract H-atom from DNA backbone leading to the damage of DNA. RSH OMe O O C MeO O O O O OH O Path A MeHN HO HO Me Path B O Neocarzinostatin Chromophore O O O Ar O O SR O OH Sugar O O O C C OH C C O O SR Ar Sugar O O O O O RS O OH RS Ar OH Ar O O O O O Sugar O Triggering pathways for the neocarzinostatin chromophore DNA H- abst ruction Sugar O O Ar RS H2O DNA diradical (Cleavage) O O H O O O RS O OH O Sugar O OH Ar H O HO O Sugar O Scheme 4. Mechanism of DNA cleavage by Neocarzinostatin (zinostatin). Joint initiative of IITs and IISc – Funded by MHRD Page 8 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics The new antibiotic N1999A2 isolated from the broth filtrate of Streptomyces sp. AJ9493, possesses a novel 9-membered ring enediyne chromophore similar to neocarzinostatin. But N1999A2 is not chromoprotein. The most interesting feature is that the stable N1999A2 exists as enediyne chromophore alone. The antibiotic N1999A2, the structure of which is given below (Figure 4.), revealed more random DNA cutting profile than neocarzinostatin chromophore. OMe Cl HO O HO HO O O OH O MeHN HO HO Me O N1999A2 Figure 4. Structure of enediyne antibiotic N1999A2. 2.2.3.2. C-1027 Chromophore C-1027 is one of the most potent antitumor antibiotic chromoproteins. It composed of an 11kDa apoprotein and a highly reactive chromophore. The C-1027 chromophore is in equilibrium with its active biradical form in the apoprotein and unlike NCS does not need nucleophiles or radicals for its activation. The p-benzyne biradical thus generated exerts its potent biological activity by abstracting hydrogen atoms from the sugar portion of double stranded DNA, which ultimately leads to oxidative cleavage of DNA (Scheme 5). Mechanism of Biological Action of C-1027 Chromophore MeO O C-1027 Chromophore O O N H MeO MeO O O O O N O H H- abstruction O DNA O N OH OH O N O OH O Cl OH O O OH OH O O Cl O O OH O O DNA diradical (Cleavage) Cl Scheme 5. Cycloaromatization process of C-1027 chromophore. Joint initiative of IITs and IISc – Funded by MHRD O O N O O Page 9 of 89 O O N H O NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.2.3.3. Kedarcidin Chromophore Kedarcidin is a new chromoprotein antitumor antibiotic that was isolated from the fermentation broth of a novel actinomycete strain. It consists of an apoprotein and a cytotoxic, highly labile, non protein chromophore. The apoprotein is water soluble while the chromophore is solvent-extractable, cytotoxic and highly unstable. As with NCS, the antitumor activity of kedarcidin is due primarily to the chromophore. The enediyne core is activated by chemical reduction (e.g. sodium borohydride) followed by spontaneous cyclization to a biradical intermediate and DNA cleavage (Scheme 6). Me Me R O O MeO Cl OMe H N NH R OH O O o Nucleophilic H Attack O RO O O Me HO Me OH O O Kedarcidin Chromophore Me OH Cycloaromatization RO Me NMe2 NMe2 OH O OH O Me HO Me Nu OH O Nu Nu O O O O Me Me N Me Me HO Me HO OH O DNA DNA Single Strand Cleavage O2 DNA Radical R O Proposed mechanism of action of the kedarcidin chromophore OH O Nu Me NMe2 RO OH O Me HO Me OH O Scheme 6. Mechanism of Biological action of Kedarcidin chromophore. Joint initiative of IITs and IISc – Funded by MHRD Page 10 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.3. Chemical Synthesis of a Few Members of Enediynes Natural Products 2.3.1. Introduction Figure 5. History and basic strategies of total synthesis of enediyne natural products. Joint initiative of IITs and IISc – Funded by MHRD Page 11 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.3.2. Total Synthesis of Neocarzinostatin Joint initiative of IITs and IISc – Funded by MHRD Page 12 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 13 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 14 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.3.3. Total Synthesis of Calicheamicin Joint initiative of IITs and IISc – Funded by MHRD Page 15 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 16 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 17 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 18 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 19 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 20 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 21 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 22 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 23 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 24 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 25 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 26 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 27 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.3.4. Total Synthesis of Dynemicin A Retrosynthetic Analysis for the synthesis of Dynemicin A Carboxylation Me N CO2TIPS (Myers) OH O H HN O Me CO2H O OMe OMe 40 (Schreiber) H H HN Me CO2H O OMe Dynemicin A OH O O H OH Intramolecular Nucleophilic Addtion (Danishefsky) 42 OH Transannular Diels-Alder Tandem Stille reaction Yamaguchi alkynylation Me N CO2MOM O OMe H Yamaguchi alkynylation Me AllocN Amidation OH 43 OTBS Yamaguchi alkynylation OMe OMe O Suzuki reaction Stille reaction 41 O Epoxidation Me Me N HN 44 45 OTBS OH Joint initiative of IITs and IISc – Funded by MHRD O Diels-Alder reaction O H OH O Yamaguchi macrolactonization Page 28 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Myers's Synthesis of Dynamicin A Me Me O Me O O 1) Pd(PPh3)4, Na2CO3 [49], 1,4-dioxane, 1000 C 2) 4-ClC6H4OH, 1800 C Me 3 steps MeO Me O EtO Me 46 MeO O TfO OMe OMe NHBOC 50 1) KHMDS CeCl3, THF 2) p-TsOH acetone H Me 3 steps AllocN TBS Me OMe OMe O OH AllocN OH OTBS Me AllocN O O OMe OMe O OTBS MgBr 52 OMe TBS 1) [52], EtMgBr THF, AllylOCOCl 2) mCPBA, DCM PH 7 buffer N N B(OH)2 49 48 47 OMenMe OMe OMenMe OH OH OTBS 54 53 55 OTBS OTMS 57 Me N 9 steps O OTMS CO2TIPS OTMS OTMS O Me CO2H MnO2, 3HF·Et3N O H OSMT H O OTMS 56 Dynemicin A OMe O OMe O H H N 58 Myers, A. G.; Tom, N. J.; Fraley, M. E.; Cohen, S. B.; Madar, D. J. J. Am. Chem. Soc. 1997, 119, 6072-6094 Danishefsky's total synthetic route to Dynemicin A Me H OH Me O CHO 4 steps O ZnCl2, CHO DCM, 25 °C H 60% OMe OMe 59 OMe 60 Me CHO AllocN O Ph OH OH OTBS OTBS 62 63 Me O Ph O Ph 12 steps Teoc Teoc Me3Sn SnMe3 N Pd(PPh3)4, DMF, 75°C O OH OH O OH OH 81% H H OTBS OTBS 2) NH4OAc HOAc 100°C Me N 80% Ph I Me TIPS AllylOCOCl, THF I O N 3 steps 80% TIPS BrMg 1) CAN, MeCN H2O 61 Me N 64 OTBS OTBS 65 66 OMOM O 1) Tf2O, py 2) DMP, DCM 3) CrCl2, THF Me Teoc N 4 steps N O 87% CO2MOM 67 O 69 Me CO2H O OMe OMe H NOMO H OH O HN O O OTBS O Me and 3 steps H O 68 OH O OH Dynemicin A Shair, M. D.; Yoon, T. Y.; Mosny, K. K.; Chou, T. C.; Danishefsky, S. J. J. Am. Chem. Soc. 1996, 118, 9509-9525. Joint initiative of IITs and IISc – Funded by MHRD Page 29 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Schreiber's total synthetic route to Dynemicin A SnBu3 1) BTSO N Br Me MeO SiMe2thexyl OMe 70 CO2H 1) TBAF, THF 2) BrCH=CHCO2Me Pd(PPh3)4, CuI 3) LiOH H2O THF SiMe2thexyl O PyBrOP, Et3 N MeO O Cl N N 2) ClCO2Me, Me TBSO OH 71 O Me MeO N O H OMe OMe H H N O O O CO2Me OMe O O H HN 75 N Br P N N CO2H OMe OMe 2) K2CO3, MeI, acetone O O H OMe Br Dynemicin A OMe OMe O OMe Br AgOTf, 4A MS Me OMe O H OMe Me O O O CO2Me 74 OMe Me N O O OMe O MeO2C N O 73 O Me Cl 72 Me 1) O COCl OMe OMe MgBr MeO Cl thexyl = PyBrOP 76 Taunton, J.; Wood, J. L.; Schreiber, S. L. J. Am. Chem. Soc. 1993, 115, 10378-10379 2.3.5. Total Synthesis of N1999A2 Retrosynthetic Analysis for the synthesis of N1999A2 Ester Formation OH O OH HO O O HO OMe Cl N1999A2 Sonogashira Coupling Intermolecular Nucleophilic Addition Transannular Reaction PO O I PO O HO P OP OP PO (Hirama) OH Sonogashira Coupling H NMe O O (Myers) O H OH Br OP PO Elimination O PO P M I O O Glaser Coupling Intramolecular Nucleophilic Addition OP O BTSO I H M PH2 PO Joint initiative of IITs and IISc – Funded by MHRD P OP OH SnBu3 P Page 30 of 89 OH NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Hirama's Total Synthetic Route to N1999A2 1) [A], Pd(PPh3)4, CuI iPr2NEt, DMF, r.t. 2) TBAF, THF 3) TESOTf, 2,6-lutidine, -78 oC TBSO TBSO 2 Steps I I TMS TMSO O O O O 1) DIBAL, DCM, -78 oC 2) DMP, Pyr, DCM, r.t. OTES TBSO 78% over all yield OPiV O TESO PivO O O O LiHMDS, CeCl3 0 OTES THF, - 30 C TBSO OH A O TBSO O OTES OTES 68% O O 1) MsCl Et3N DMAP 2) TBAF, THF, -15 oC HO 63% TMSO OH TESO OH TMSO O O OTBS O Cl B OH CO2H DCM, -85 C 2) Et3N, DMAP, Ms2O HO OH 2) TFA, THF, H2O MeO OH OH 1) TESOTf, 2,6-lutidine 1) [B], DCC, THF HO TBSO Cl OMe HO OH O O O 3) TFA, THF, H2O, 30C 35% HO Cl OMe N1999A2 54% Kobayashi, S.; Ashizawa, S.; Takahashi, Y.; Sugiura, Y.; Nagaoka, M.; Lear, M. J.; Hirama, M. J. Am. Chem. Soc. 2001, 123, 11294. Joint initiative of IITs and IISc – Funded by MHRD Page 31 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Myers' Total Synthetic Route to N1999A2 O THPO TES LiHMDS, 1) n-BuLi, Bu3SnCl, THF 2) Cp2Zr(H)Cl, THF, Then I2, THF H HO OTHP 5 Steps TBSO BF3-OEt2 TES TES 2-Steps TMS PPh3 TBS n-BuLi, LiI O O 1) (DHQD)2PYR, K2OsO4, 2H2O K3Fe(CN)6, K2CO3, MsNH2, t-BuOH 2) FeCl3.6H2O, MeCN 3)[B], CSA, THF Br 1) O O MeS B Benzene 64% Then t-BuLi Then HOAc Mes O OH OH SnBu3 TES 1) TBAF, THF 2) DEIPSCl, TBSO Imidazole 3) NBS, DCM 72% SnBu3 H TBS OSiEt2(i-Pr) LiHMDS, THF-Toluene H H O O TBS OMe H Mes O O Cu(OAC) 2 THF-Pyr OH H OH Mes OSiEt2(i-Pr) O O OH 75% Br OSiEt2(i-Pr) H MeS H MeS O O OTES 1) Et3N.3HF, MeCN, 2)[C], DCC, THF. O O OH O OH 3)TBAF, O-NO2-Phenol 4) TESCl, Et3N OSiEt2(i-Pr) MeS OH OH OMe TBS O O 26% 2) K2CO3, MeOH O H H O O H TBSO [A], Pd(PPh3)4, CuI, Et3N, SnBu3 A TES O I TBSO OH (i-Pr)Et2SiO 30% Cl OMe OH HO OH OH O 1) TsCl, DANCO O 2) TFA, THF/H2O HO Cl OMe N1999A2 CO2H O (i-Pr)Et2SiO C Cl OMe Ji, N.; O-fDowd, H.; Rosen, B. M.; Myers, A.G. J. Am. Chem. Soc. 2006, 128, 14825-14827 Joint initiative of IITs and IISc – Funded by MHRD Page 32 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.3.6. Total Synthesis of Kedarcidin Chromophore Joint initiative of IITs and IISc – Funded by MHRD Page 33 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 34 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.3.6.1. Approaches for the Synthesis of Kedarcidin Chromophore Aglycon Two Approaches for the Synthesis of Kedarcidin Chromophore Aglycon i-PrO OH O CH3O CH3O HN O N O Cl OR2 O Kedarcidin Chromophore R1O O Aldehyde Addition Cyclisation Transannular Cyclisation BocHN ArHN O N O Cl N OPMBM Cl O O OTIPS OTES O OH O TBSO Br CHO OTBS TBSO H M. Hirama 2007 (partial synthesis) A. G. Myers 2002/2007 Hirama‘s Synthesis of Kedarcidin Chromophore Aglycon 2.3.6.2. Hirama’s Retrosynthetic Analysis of Kedarcidin Chromophore Aglycon i-PrO i-PrO i-PrO OH O CH3O CH3O N O CH3O CH3O HN Cl OMOM O Amide Formation O Cl O Cl OPMBM OPMBM OTES O O Dehydration Sonogashira TBSO O TMS OH RO O HO O N OTMS O CH3O BocHN O N OR' O CH3O HN O OMOM TBSO Cyclisation O TES O Epoxide Formation BocHN Ester Formation BocHN OH N O Cl N HO OMOM HO H OH O I TES O O Joint initiative of IITs and IISc – Funded by MHRD O OPMBM O Cl OTES O Ether Bond Formation TBSO H I TES O O Page 35 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Hirama‘s Synthesis of Kedarcidin: Part-1: Synthesis of Naphthalene Aromatic Core 2.3.6.2.1. Part-1: Synthesis of Naphthalene Aromatic Core (Hirama's Synthesis) HO CO2Me HO Ph2CCl2 175oC O HO Ph OH i-PrO H3CO CO2Me 1. i-PrI, K2CO3, acetone, 50oC 2. AcOH, H2O, reflux, 72% (3 steps) O Ph O CO2Me 1. NaOH, MeOH, i-PrO H2O, 60oC Br 2. (COCl)2, PhCl3 H3CO OCH3 Br CO2Me 1. NBS, THF, r.t. 2. MeI, K2CO3, i-PrO Acetone, 50oC 86% (2 steps) HO OH CH2N2, Et3N, Et2O, Cl then PhCO2Ag, Et3N, i-PrO MeOH, 75% (3 steps) H3CO OCH3 CH2=CHCO2t-Bu, Pd(OAc)2, P(p-tol)3, Et3N, 100oC, 90% i-Pr2NEt, PhMe, r.t., 62% (3 steps) i-PrO CO2Me H3CO CO2t-Bu 1. Ba(OH)2.8H2O, t-BuOH 2. (COCl)2, DCM CH3O Br OCH3 i-PrO COCl H3CO CO2t-Bu OCH3 OCH3 i-PrO CO2Me OH TFA i-PrO CO2t-Bu 99% CH3O OCH3 Joint initiative of IITs and IISc – Funded by MHRD OH CO2H OCH3 A Page 36 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 37 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.3.6.2.2. Hirama‘s Synthesis of Kedarcidin: Part-2: Synthesis of β-Amino Acid Core 2.3.6.2.3. Hirama‘s Synthesis of Kedarcidin: Part-3: Synthesis of Fused 5-Membered Ring System of Enediyne Core Part-3: Synthesis of Fused Ring of Enediyne Core (Hirama's Synthesis) OMe MeO 1. MeO O HO OH PMB MeO O X Amberlyst-15 OH 2. MOMCl 3. PPTS/MeOH 72% (3 steps) OH MOMO OH OMOM X = OH X=I O OMOM MOMO i-PrMgCl, CH2I2 I 2. I2, Pyridine 89% (2 steps) MOMO O -78 to 0oC, THF 69-71% 1. (Cl3C)2C=O, Pyridine TBSO 2. PPTS, 2-butanone 3. TBSCl, imidazole 77% (3 steps) TBSO I O Joint initiative of IITs and IISc – Funded by MHRD CH2Cl2, r.t. 92% OH OMOM MOMO HClO4 I O MOMO OMOM MOMO OH THF(aq) 61% OMOM MOMO I MOMO HO OH HO 3. TBAF, THF, 0oC HO O MOMO 1 (0.2 mol%) 1. DIBAL-H, -80oC 2. (MeO)MeC=CH2 O O I2/PPh3 99% MOMO THF, r.t., 96% (2 steps) OH HO EtOH (aq), reflux Ph3P=CH2, MOMO 1. TPAP, NMO Zn dust I O O DIAD, PPh3, THF O 72% (4 steps) I O O O (C) Page 38 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Grignard via Iodine-Magnesium Exchange THF CH2I2 i-PrMgCl i-PrI ClMg I O MOMO I ClMg I O O MOMO O O I O MOMO MOMO I I MOMO MOMO O - O I Hirama‘s Synthesis of Kedarcidin: Part-4: Synthesis of Enediyne Core 2.3.6.2.4. Part-4: Synthesis of Enediyne Core (Hirama's Synthesis) O O O HO OH O O 1) NaIO4, SiO2, DCM, H2O 2) H TMS n-BuLi, THF, -78°C TMS 74% (2 steps) PDC, MS 3A, OH O O MgBr O O H TMS O OH Et2O, -78°C O 63% (2 steps) DCM TMS H O 1) n-BuLi, THF, -100°C, then, TESCl, -80°C, 80% 2) LiOH, THF, H2O, 97% H O HO 1) TFA, THF, H2O (2 : 10 : 5), 93% H OTES Joint initiative of IITs and IISc – Funded by MHRD PMBMO 1) PMBMCl, i-Pr2NEt,DCM, 76% OH H TES OH OH 2) TFA, THF, H2O (1 : 20 : 10), 90% 2) TBSCl, DMAP, Et3N,DCM, > 99% TES OTBS TES Page 39 of 89 (D) NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Hirama‘s Synthesis of Kedarcidin: Part-5: Attaching A-D Units with Enediyne Core 2.3.6.2.5. Part-5: Attaching A-D Units with Enediyne Core (Hirama's Synthesis) O C BocHN 1. 2, CsF DMF, 60 oC I BocHN N O O O Cl 2. TBSCl 90% (2 steps) 1. 0.3 M KOH(aq) OMe O + CO2Me I PMBMO OPMBM O Cl OTES O 3.D, TESCl, Imidazole, DMF, 95% TBSO BocHN O N 2. 3, EDC.HCl, DMAP 86% (2 steps) H I TBSO OH O TES O O H N O OH B Cl OH D TES Pd2(dba)3. CHCl3(0.5 equiv.) CuI (2 equiv.), i-Pr2NEt (36 equiv.), Cl i-PrO BocHN O N O OPMBM OTES O DMF (0.001M), r.t., 1h, (44-47%) TBSO O TES O 1. TBSOTf, 2-6-lutidine, DCM OH O CH3O CH3O 2. SiO2, DCM 3. A, HOAt, EDC.HCl, DCM, 89% (3 steps) i-PrO HN CO2H TBSO O O OTES O R1 O N N N N .HCl OH O TES N EDC.HCl O OPMBM O TBSO N C N O N O TES O Peptide Coupling HN Cl OTES A OAllyl CH3O AllylBr, Cs2CO3, CH3O DMF, 0°C, > 99% OPMBM O CH3O i-PrO O Cl OH CH3O O N N P.T. R1 R1 O R2 NH2 O O R1 N HO O HN O N N N N N N H N P.T. -HOAt R1 HN R2 Joint initiative of IITs and IISc – Funded by MHRD N Page 40 of 89 O N H R2 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Hirama‘s Synthesis of Kedarcidin: Synthesis of Cyclic Enediyne Core: Partial Synthesis of Kedercidin 2.3.6.2.6. Part-6: Synthesis of Cyclic Enediyne Core: Partial Synthesis of Kedercidin i-PrO OAllyl CH3O O CH3O TBAF, THF HN OAllyl CH3O O CH3O 0°C, 93% HN O N OTES OPMBM OH O HO O TES O O H i-PrO i-PrO OAllyl OAllyl 1. TFA, THF, H2O CH O (2 : 10 : 5), 50°C, 71% 3 CH3O 2. TBSCl, Et3N, DMAP, DCE, 85% O Cl OPMBM O O O N O Cl TBSO i-PrO O CH3O 1. IBX, MS 4A DCM-DMSO (10 : 3) CH3O O HN 2. TMSOTf, 2,6-lutidine, DCM, -70°C O N O Cl N Cl OPMBM O O OPMBM OTMS O OH O HN CHO TBSO HO TBSO OH H Unstable, use immediately H i-PrO OAllyl CH3O O Additive 30-50 equiv. CH3O YbCl3-LHMDS 30-50 equiv. THF [1 mM], -25°C, 25h 22% (3 steps), 2/3 ( i-PrO N Cl OAllyl O H3CO HN THF [1 mM] CH3O HN O O OTMS OPMBM OTMS O TBSO O TMS OH N + Cl Kedarcidin O O OPMBM OTMS O TBSO O TMS OH Key steps: (a) Nucleophilic Addition-Cyclisation; (b)1% overall yield from the longest linear sequence (16 steps); (c)Still 4 steps to achieve the total synthesis of kedarcidin chromophore aglycon Joint initiative of IITs and IISc – Funded by MHRD Page 41 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Myers‘ Synthesis of Kedarcidin Chromophore Aglycon 2.3.6.3. Myers’ Retrosynthetic Analysis for the Synthesis of Kedercidine i-PrO OH O CH3O CH3O i-PrO OMOM i-PrO CH3O O CH3O CH3O HN OR' HN O N O O CH3O HN O N Cl OMOM O N O Cl OTIPS O Cl OTIPS OH O OH O Dehydration O Br RO O HO TBSO Transannular Cyclisation Epoxidation Glaser Cyclisation i-PrO BocHN O Cl O S O Ether Formation CH3O O Br Ester Formation HN HO O O Cl HO O O H O TBS OTIPS OH H Br Amide Formation O CH3O N TBSO Br O O O Cl HO O CH3O OMO CH3O OCH3 N Br OMOM BocHN OCH3 N i-PrO Br TBSO TBS Pd Reation H TES Myers‘ Synthesis of Kedarcidin: Part-1: Synthesis of Naphthalene Aromatic Core 2.3.6.3.1. Part-1: Synthesis of Naphthalene Aromatic Core (Myers' Synthesis) HO i) PPh3, DEAD, i-PrO i-PrOH, THF O ii) NaOH, H2O, 93% O O 1) CH(OEt)3, SnCl4, DCM HO OH i-PrO 2) CH3I, K2CO3, DMF H3CO 79% (2 steps) i) (CH3)2NCH2CH2N(CH3)Li, THF, -20°C CHO OCH3 O i-PrO H3CO CH3 i) (CH3)2NCH2CH2N(CH3)Li, i-PrO THF, -20°C CHO ii) n-BuLi, -20°C OCH3 iii) CO2, 88% i-Pr2NEt, PhH, r.t. i-PrO 89% (2 steps) CH3O H3CO OH 1) EtO CO2H THF, 96% CHO OCH3 NaOH, EtOH/H2O, CO2Et OCH3 Joint initiative of IITs and IISc – Funded by MHRD 60°C, 99% Li ii) n-BuLi, -20°C iii) CH3I, 92% O P OEt OEt 2) (COCl)2, PhH i-PrO COCl CO2Et H3CO OCH3 i-PrO OH CH3O CO2H OCH3 A Page 42 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Myers‘ Synthesis of Kedarcidin: Part-2: Synthesis of β-Amino Acid Core 2.3.6.3.2. Staudinger Reaction R-N3 PX3 R-N-N=N-PX3 -N2 phosphazide NN+ N Cl MOMO N Cl O OCH3 aza-ylide PPh3 N N N PPh3 Cl N H2O R-N=PX3 - X3P=O N N N PPh3 R-NH2 PPh3 N -N2 Cl MOMO MOMO O OCH3 Joint initiative of IITs and IISc – Funded by MHRD MOMO N N O O OCH3 Page 43 of 89 OCH3 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.3.6.3.3. Myers‘ Synthesis of Kedarcidin: Part-3: Joint initiative of IITs and IISc – Funded by MHRD Page 44 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 45 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.3.6.3.4. Myers‘ Synthesis of Kedarcidin: Part-4: Synthesis of Diyne Core Joint initiative of IITs and IISc – Funded by MHRD Page 46 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Myers‘ Synthesis of Kedarcidin: Synthesis of Cyclic Enediyne Core 2.3.6.3.5. i-PrO i-PrO OH O CH3O CH3O CH3O 1) [VO(acac)2] (0.2 equiv.), CH3CPh2OOH (1.3 equiv.), PhH O O Cl O CH3O HN N OH OTIPS O OTIPS OH O TESO O i-PrO Martin Sulfurane Dehydration CH3O PhH, 83% O Cl HO Martin Sulfurane Dehydration CF3 F3C Ph O Ph S Ph O Ph CF3 F3C (10 equiv.), O N 2) TESCl (50 equiv.), Imidazole (100 equiv.), DCM, 32% (2 steps) OH HN CH3O OH HN Cl R1 R1 R 2 F3C Ph O S Ph O Ph R3 F3C OH R2 R3 Martin Sulfurane N F3C HO O TESO O CF3 F3C Ph O Ph S Ph O Ph CF3 F3C O O Ph CF3 O OTIPS Joint initiative of IITs and IISc – Funded by MHRD Ph O S F3C Ph HO Ph CF3 R1 R2 Page 47 of 89 R3 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.3.6.3.6. Overview Myers‘ Synthesis Joint initiative of IITs and IISc – Funded by MHRD Page 48 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.3.6.4. Summary of Kedarcidin Synthesis Summary of Kedarcidin Synthesis Hirama’s Synthesis 16 steps, 1% overall yield Still 4 steps to go Key Step: Aldehyde Addition Cyclisation i-PrO OH O CH3O CH3O HN O O N O Cl H3CN(CH3)2 OH O O H3C HO H3C OH O O O Kedarcidin Myers’ Synthesis 25 steps, 1% overall yield Synthesis applied to Kedarcidin glycon in 2007 Key Step: Transannular Cyclisation Joint initiative of IITs and IISc – Funded by MHRD Page 49 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.3.7. Total Synthesis of Maduropeptin Joint initiative of IITs and IISc – Funded by MHRD Page 50 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 51 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 52 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 53 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 54 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 55 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Joint initiative of IITs and IISc – Funded by MHRD Page 56 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.4. Biosynthesis of a Few Members of Natural Enediynes 2.4.1. Introduction Microorganisms mostly from soil and marine natural source, (such as bacteria, fungi) produce a large variety of substances (secondary metabolites) with a vast diversity of fascinating molecular architecture and potent biological activities that are not available in any other systems. Enediyne class of natural product is of such fascinating examples produced by microorganisms and isolated. Biosynthesis of secondary metabolites includes (a) finding the reactions available in nature, (b) study of the enzymatic mechanisms of these reactions, (c) investigating how these reactions are linked to produce complex architechture, and (d) the regulatory mechanisms of the pathways they are formed, (e) to manipulate nature's biosynthetic machinery for the discovery and development of new drugs of microbes origin. The biosynthesis of the enediynes is intriguing because of the uniqueness of the chemical structures of these classes of molecules. The origin of the enediyne core was initially studied using isotope-labeling experiments (by monitoring the production of neocarzinostatin, dynemicin, and esperamicin) which established the acetate as a precursor unit. However, the answer of whether the enediyne core was constructed by the degradation of fatty acids or by de novo biosynthesis with a dedicated fatty acid or polyketide synthase (PKS) was unclear. Cloning and characterization of the gene clusters for 5 enediynes, [C-1027, neocarzinostatin (NCS), calicheamicin, dynemicin, and maduropeptin], led the foundation of investigating enediyne biosynthesis. Since then molecular and biochemical studies are being undertaken for deciphering enediyne biosynthesis. Joint initiative of IITs and IISc – Funded by MHRD Page 57 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.4.2. Enediyne Biosynthesis in the Pre-Genomic Era In early 1988, Schreiber and Kiessling devised concised synthetic routes for the enediyne cores of calicheamicins and neocarzinostatin. They envisaged that the branched molecule A might be the common biosynthetic precursor for both the 9- and 10-membered enediynes. Considering this, they demonstrated that a series of steps including (a) electrocyclic ring closure, (b) proton transfer and (c) oxidation could transform A into a 9-membered enediyne (pathway I, Figure 6). On the other hand an intra-molecular Diels–Alder reaction followed by the addition of one carbon unit at the acetylene terminus could convert structure A into a 10-membered enediyne (pathway II, Figure 6). Biosynthetic Mechanism Postulated for Enediynes in early 1988, by Schreiber and Kiessling Path I 9-Membered Ring Enediyne core A Path II OR 9-Membered Ring Enediyne core Figure 6. An early biosynthetic mechanism postulated for enediynes. Joint initiative of IITs and IISc – Funded by MHRD Page 58 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Later on, in 1989, Hensens et al. produced 13C-enriched neocarzinostatin by feeding 13Clabeled acetate to Streptomyces carzinostaticus. This study confirmed the structures of neocarzinostatin chromophores proposed by Edo and Myers. The first clue about the biosynthetic origins of the molecular building blocks also came from their studies. The experiment with singly labeled acetate suggested that the bicyclo[7.3.0]dodecadienediyne moiety could be derived from a linear precursor that consists of seven acetate units (Figure 7). That the acetates are assembled in a head-to-tail fashion was evident from the study of culturing with doubly- and mixed-labeled acetate. Most importantly, they envisaged that the two carbons of the -yne group originate from the same acetate unit, which brings the difference between the 9- and 10membered enediynes. They also proposed that the linear precursor derived from oleate or crepenynate is truncated on both ends prior to cyclization. Subsequent oxidation, cyclization and oxygenation would furnish the fully functionalized 14-carbon enediyne core. Folding Pattern for the 9-Membered Enediyne Core of Neocarzinostatin 10 11 9 8 7 O 1 2 3 4 6 OR 12 CH3COOH 5 14 13 9-Membered Ring Enediyne Core Figure 7. Folding pattern for the 9-membered enediyne of neocarzinostatin. Joint initiative of IITs and IISc – Funded by MHRD Page 59 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Another isotopic labeling study by Tokiwa et al. revealed a very different 13C-incoporation pattern for the bicyclo[7.3.1]-tridecadiynene core of dynemicin A isolated from Micromonospora chersina M956-1. They proposed that the bicyclic enediyne core and the anthraquinone moiety of dynemicin A are derived from two heptaketide chains that consist of two sets of seven acetate units. There are two possible pathways for the linear heptaketide to fold into the final bicyclic structure (Figure 8). Identification of the incorporated acetate units from the [1,2-13C2] acetate feeding experiment suggested that the cyclization followed pathway (a) rather than path (b). From their results, it was also clear that the two carbons of one -yne group in dynemicin are derived from different acetate units which is contrary to the early finding that the two carbons of the -yne group in neocarzinostatin originate from the same acetate unit. From the earlier studies it was postulated that uncialamycin, the other member of the dynemicin-type enediyne subfamily containing a different enediyne core, is derived from a dynemicin-like precursor. Folding Pattern for the 10-Membered Enediyne Core of Dynamicin A 13 11 10 9 12 (a) 14 1 8 2 (b) 7 3 6 4 5 CH3COOH 10-Membered Ring Enediyne Core Figure 8. Folding patterns for the 10-membered enediyne of dynemicin A. Joint initiative of IITs and IISc – Funded by MHRD Page 60 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics A similar isotopic labeling experiment was performed for investigating the biosynthetic route to the esperamicin producing Actinomadura verrucosospora. This experiment suggested that the enediyne of esperamicin A1 is also assembled from seven acetate units in a head-to-tail fashion. In this case, there four possible pathways were proposed for the linear heptaketide to fold into the bicyclic structure (Figure 9). Identification of the incorporated acetate units, particularly the starting acetate unit C11–C12, ruled out the pathways a, b and d. It was observed that the production of esperamicin A1 by A. verrucosospora was significantly reduced by administrating cerulenin, an inhibitor that targets the β-ketosynthase (KS) domain of both fatty acid synthase (FAS) and polyketide synthase (PKS), whereas supplementation of the culture with the fatty acid oleate did not restore the biosynthesis. These observations suggested that the enediyne core is probably derived from a polyketide precursor, rather than a fatty acid one. Further, the three isotopic labeling experiments provided clues that linear polyketide precursors consist of head-totail coupled acetate units. The differences in isotope incorporation pattern among neocarzinostatin, calicheamicin and dynemicin indicated the different origins of the -yne carbons and hence suggesting the different biosynthetic pathways for these enediynes. Folding Pattern for the 10-Membered Enediyne Core of Esperamicin A1 14 11 15 11 10 9 12 13 1 14 2 (a) (b) 14 6 4 11 5 15 13 11 10 9 12 13 1 4 6 4 5 6 4 5 14 (d) CH3COOH 11 15 10 9 13 1 8 2 7 3 6 12 8 2 3 7 3 (c) 7 8 2 15 8 2 10 9 12 1 14 13 1 7 3 10 9 12 8 15 10-Membered Ring Enediyne Core 5 7 3 6 4 5 Figure 9. Proposed biosynthetic pathways for the linear heptaketide to fold into the bicyclic structure of 10-membered ring enediynes. Joint initiative of IITs and IISc – Funded by MHRD Page 61 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.4.3. Enediyne Biosynthesis in the Genomic Era As was revealed from the isotopic labeling experiments, little new insight was gained on the biosynthesis of the enediynes. Therefore, in the genomic era effort were undertaken to locate the putative type I or type II PKS gene on the genomic DNA of the enediyne producers (microorganisms) by DNA hybridization. However these were unsuccessful. Instead, Shen and coworkers succeeded in locating the DNA fragment that encompasses the enediyne gene clusters for C-1027. They were able to find out the gene (cagA) that encodes the chromophore-associated apoprotein CagA and two putatively conserved deoxysugar synthesizing genes. At the same time, Thorson et al. were able to identify the gene locus for calicheamicin biosynthesis. In 2002, the completion of the sequencing and partial annotation of the C-1027 and calicheamicin gene clusters was announced. Since then the study of enediyne biosynthesis entered the genomic era. Therefore, genetic manipulations of genes related to secondary metabolism now offer a promising tool to investigate the biosynthetic pathway of formation of and prepare complex natural products like enediyne biosynthetically. This approach depends on the cloning and genetic and biochemical characterization of the biosynthetic pathways of the metabolites. Thus, several research groups are involved in the cloning, sequencing, and characterization of the several enediyne biosynthesis gene clusters from the producer microorganisms through which the convergent biosynthetic strategies for C-1027, NCS and other enediynes were developed. Manipulation of genes governing enediynes biosynthesis allowed one to engineer enediyne compounds. This approach offers the opportunity to decode the genetic and biochemical basis for the biosynthesis of enediynes and many other structurally complex natural products and to explore ways to make more antitumor agents. Here are few examples of biosynthesis of enediynes 2.4.3.1. The Apoprotein and The Gene Cluster for Enediyne Biosynthesis The Apoprotein: All known 10-membered enediyne natural products were isolated as freestanding chromophores. On the contrary, the nine-membered enediynes were isolated as a chromoprotein complex- a binding protein known as protective apoprotein covering the dissociable enediyne chromophore. Exception is N1999A2 where no apoprotein was found. The 9-memred enediyne chromophores are extremely unstable in aqueous solution. For example neocarzinostatin is extremely unstable in aqueous solution in the absence of the apoprotein but the apoprotein cover greatly enhances the stability of the labile chromophore. However the apoproteins are not similar for all the enediynes. As for example, the maduropeptin apoprotein (MdpA) does not share similarity with the apoproteins of neocarzinostatin, C-1027 and kedarcidin. The 133 amino acid long MdpA represents a new protein class that does not share significant sequence homology with any protein deposited in the National Center for Biotechnology Information (NCBI) database. The structures of neocarzinostatin and C-1027 associated with their apoproteins (NcsA and CagA) have been determined which reveals that the apoproteins share an immunoglobulin-like fold that consists of a seven-stranded antiparallel β-barrel and two additional β-strands. The βstrands and the three loops of the apoprotein forms a hydrophobic pocket in which the Joint initiative of IITs and IISc – Funded by MHRD Page 62 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics chromophore (the reactive enediyne core) is accommodated. The structure of the protein– neocarzinostatin complex revealed that the reactive sites of the cromophore such as the epoxide, the acetylene groups, and the nucleophilic addition site are greatly shielded from the solvent. Similarly structure determination of the aromatized C-1027 chromophore revealed that the enediyne core and the the hydrophobic benzoxazolinate moiety interact and reside on the hydrophobic residues of the protein. These bindings bring stability of the extremely reactive 9membered enediynes. Role of Binding Proteins: The binding protein directs transport of the reactive enediyne chromophore to the extracellular environment. It is also found to be essential for self-resistance by stabilizing the reactive enediyne chromophore. The 9-membered enediynes C-1027 and maduropeptin readily undergo cycloaromatization in the absence of the binding protein. Thus the binding proteins help stabilizing the enediyne in their native form. Establishment of the amino acid sequence of the homologous binding proteins enables the cloning and the sequencing of the genes-cagA for C-1027 and ncsA for NCS. Since there is no binding protein for the ten-membered enediyne calicheamicin, a different strategy was used to clone the gene cluster of ten-membered enediyne calicheamicin. The strategy utilized to clone and localized the gene cluster of ten-membered enediyne calicheamicin are-(a) screening of clones that are capable of conferring calicheamicin resistance using PCRbased screens and (b) followed up by DNA-shotgun sequencing. 2.4.3.2. General Biosynthesis of Enediyne Cores Though there exists various types of gene clusters for different enediynes, a unified biosynthetic scheme for the nine- and ten-membered enediyne cores is now possible owing to the discovery of a shared iterative type I PKS (polyketide synthase). Bacterial polyketide biosynthesis carries out either by noniterative, modular type I PKS, or a multienzyme complex of iteratively acting type II PKS, or homodimeric, iteratively acting condensing type III PKS without an acyl carrier protein (ACP). In contrast, the enediyne PKSs from the biosynthetic pathways of calicheamicin 1I and C1027 seem to resemble a family of fungal iterative type I PKSs. To date, five gene clusters cloned only one type of PKS that are all iterative type I PKS. The gene cluster for C-1027 contains a single PKS. This PKS is shared by sequence homology and domain architecture among the enediyne family (Fig. 4A). Disruption of this SgcE (the PKS for C-1027 synthesis) in the producer of C-1027 did not produce C-1027. Identical results were observed with the homologous neocarzinostatin PKS, NcsE. These results provide strong support that the enediyne core is produced by a polyketide pathway. The enediyne PKSs use a single set of catalytic domains for polyketide synthesis. Bioinformatic analysis of this PKS family, PKSE, showed that the enediyne PKS encompass four domains in the following order (N- to C-terminus): (a) a ketosynthase (KS), (b) an acyltransferase (AT), (c) a ketoreductase (KS), and (d) dehydrogenase (DH). Enediyne PKS possess closest sequence homology to polyunsaturated fatty acid (PUFA) synthases involved in the biosynthesis of docosahexaenoic acid in Moritella marina and eicosapentaenoic acid in Shewanella (Figure 10). The significant sequence identity and Joint initiative of IITs and IISc – Funded by MHRD Page 63 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics similarity shared by PKSs, CalE8 (the PKS for calicheamicin synthesis) and SgcE (the PKS for C-1027 synthesis) indicated that the 9- and 10-membered enediyne cores may share a common PKS progenitor. Thus, the iterative type I PKS is unique to all of the enediyne family. It is now evident that the enediyne biosynthetic gene clusters share conserved architecture featuring the enediyne PKS. These enediyne PKS formed the basis of several expedient strategies to clone additional enediyne biosynthetic gene clusters such as of NCS, maduropeptin, and dynemicin gene clusters via a PCR approach. All the studies towards polyketide biosynthesis by enediyne PKSs established that with selfphosphopanetheinylation of a unique ACP, PKSE catalyses the enediyne synthesis. Therefore, enediyne biosynthesis follows an ACP-dependent PKSE-catalyzed pathway (Figure 11). Architecture and domains of the enediyne PKSE and its relationship with PUFA synthase Figure 10. Architecture and domains of the enediyne PKSE and its relationship with PUFA synthase (Lanen, S. G. V.; Shen, B. Curr Top Med Chem. 2008, 8, 448–459.). Joint initiative of IITs and IISc – Funded by MHRD Page 64 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Proposed pathway for biosynthesis of putative nine- or ten-membered enediyne cores by enediyne PKS Enediyne PKS Accessary Enzymes (9-membered specific) 9-Membered Enediyne O 9-Membered Enediyne Core (Neocarzinostatin) SCoA Me ACP Acetyl-CoA PKSE R + O O O OH SCoA Malony-CoA (7 x) CO2 Polyketide Intermediate (7 x) Enediyne PKS Accessary Enzymes (10-membered specific) (Dynamicin A (Esperamicin Core) Core) 10-Membered Enediyne Core 10-Membered Enediyne Figure 11. Proposed pathway for biosynthesis of a polyunsaturated intermediate from acyl CoA by PKSE and the subsequent transformation by enediyne PKS associated enzymes into putative nine- or ten-membered enediyne cores that are finally tailored to individual enediyne natural product. Atoms that were incorporated intact from acyl CoA precursors to the enediyne cores are shown in bold. 2.4.3.3. Peripheral Moieties of the Enediyne Chromophores As was discussed that the enediyne first bind to the DNA sequence specifically and then breaks the double strand or single strand DNA via the generation of enediyne biradical through the well known BC or MSC that abstract –H from the sugar phosphate backbone of DNA. Now the sequence specific binding of enediyne to DNA and the physical properties of the enediyne chromophores are largely depend on the peripheral moieties encompassing the enediyne warheads. The peripheral moieties or building blocks can be divided into two groups- (a) aromatic and (b) sugar moieties. The aromatic moieties are essential for DNA–chromophore interaction through intercalation. The aromatic unit benzoxazolinate present in C-1027 and orsellenic acid in calicheamicin are found to intercalate into DNA. Besides these aromatic units, many enediyne chromophores contain mono- or polysaccharides that play important roles in fine-tuning the interaction between the chromophore and DNA. Joint initiative of IITs and IISc – Funded by MHRD Page 65 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Figure 12. Building blocks of enediyne chromophores. The sites on the enediyne cores for peripheral moiety attachment are indicated by the big dots. Nature has created the structurally diverse enediyne chromophores, 13 numbers till the date by using a small collection of aromatic and sugar moieties (Figure 12). Connecting these moieties at different positions of the enediyne cores contributes to the structural diversity of enediyne chromophores. More of such aromatic and sugar moieties are expected to discover that will help deciphering new enediyne chromophores in the future. Joint initiative of IITs and IISc – Funded by MHRD Page 66 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Success of the sequencing and annotation of the enediyne gene clusters and corroboration of the biosynthetic pathways has discovered a number of novel enzymes and chemical transformations in the biosynthesis of the aromatic and sugar moieties and the enediyne cores. These pathways and enzymatic transformations to the aromatic and sugar moieties as well as enediyne cores are given below (Figure 13). Also the successful stories of exploiting the biosynthetic pathways for producing C-1027 and calicheamicin 1I are highlighted. O SCoA Me Acetyl-CoA PKSE AviM (for Avilamycin) or CalO5 (for Calicheamicin) Enz S + O Enz S O Me O Me O Avilamycin O OH OH O OH Orsellinic acid O CO2 SCoA Calicheamicin) (3 x) Malony-CoA (3 x) O SCoA Me Acetyl-CoA O PKSE NcsB (for Neocarzinostatin) + O O O OH S Enz O O O Me O O HO NcsB S Enz O O O OH Me OH Me Naphthoic acid (5 x) Malony-CoA (5 x) Enz S O MeO OH NcsB3 O NcsB CO2 SCoA Enz S Neocarzinostatin NcsB1 Me Naphthoic acid Figure 13. General biosynthesis of peripheral aromatic moieties of enediynes. Joint initiative of IITs and IISc – Funded by MHRD Page 67 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.4.4. Biosynthesis of C-1027 The C-1027 chromophore can be dissected into 4 biosynthetic building blocks which are (a) an enediyne core synthesized via polyketide pathway by enediyne PKSs. (b) a β-amino acid annotation of the gene products of the C-1027 gene cluster suggested that the β-amino acid is derived from L-tyrosine. (c) a deoxy aminosugar initial annotation of the gene products of the C-1027 gene cluster suggested that the deoxy amino sugar is derived glucose-1-phosphate. (d) a benzoxazolinate moiety The biosynthesis of the benzoxazolinate moiety was originally proposed to start from anthranilate, a common intermediate from the shikimate pathway. Subsequent characterization of the enzymes from the putative pathway revealed an unexpected pathway with chorismic acid as the starting material (Figure 14). Model of Convergent Biosynthesis of C-1027 O SCoA H3C OH H2C Acyl Transferase + O OH Benzoxazollinate O SCoA Acetyl-& Malony CoA O MeO O O Enediyne Core O O O OH O O Me2N H3C HO HO OH O OH HO OP Deoxy Aminosugar D-Glucose- 1-Phosphate Glycosyl transferase O HO OH Chorismic Acid PKSE N H OH O O O NH2 O NH2 Cl Condesation Enzyme OH -Amino Acid L-Tyrosine Figure 14. A model of convergent biosynthesis for the enediyne C-1027. Sequencing of the gene cluster revealed homologs for a glycosyl transferase, an acyltransferase, and condensation enzymes, suggesting a convergent biosynthetic approach is used in enediyne assembly for C-1027. The initial enzymatic steps in every biosynthetic pathway for C-1027 have been analyzed using a combination of in vivo gene inactivation and in vitro characterization of recombinant enzymes. This experiment established the starting metabolite for each moiety and provided substantial evidence for a convergent approach in enediyne biosynthesis. Joint initiative of IITs and IISc – Funded by MHRD Page 68 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Enediyne core (4) biosynthesis: There are 13 genes identified within the C-1027 gene cluster that are consistent with the structure of C-1027 and are essential for the production of enediyne core of C-1027. These are sgcE, sgcE1, sgcE2, sgcE3, sgcE4, sgcE5, sgcE6, sgcE7, sgcE8, sgcE9, sgcE10, sgcE11 and sgcF that encode the enediyne core (4) biosynthesis (Figure 15). Figure 15. Organization of the C-1027 biosynthesis gene cluster ORFs outside the sgcB1 to sgcR3 region are not essential for C-1027 production. Color coding indicates the biosynthesis genes for the enediyne core (red), deoxyaminosugar (blue), β-amino acid (green), benzoxazolinate (purple), and all other genes (black). (Liu, W.; Christenson, S. D.; Standage, S.; Shen, B.Science 2002, 297, 1170.) It is not clear whether the enediyne cores are assembled by de novo polyketide biosynthesis or degradation from a fatty acid precursor. However, feeding experiments with 13C-labeled precursors supported that both the nine- and 10-membered enediyne cores are derived from a minimum of eight acetate units organized in head-to-tail. Importantly, only one gene, sgcE, among all other genes indentified within the C-1027 cluster encodes a PKS. SgcE contains five domains—(a) the ketoacyl synthase (KS), (b) acyltransferase (AT), (c) ketoreductase (KR), (d) dehydratase domains that are characteristic of known PKSs, and (e) a domain at the COOHterminus that is unique to enediyne PKSs. Moreover a putative acyl carrier protein domain is proposed to present in between AT and KR region (Figure 16). Most importantly, the SgcE enediyne PKS exhibits head-to-tail sequence homology. It has an identical domain organization to the CalE8 enediyene PKS that catalyzes the biosynthesis of the 10-membered endiyne core of calicheamicin in Micromonosporaechinospora. Therefore, the nine- and 10-membered enediyne cores share a common polyketide biosynthetic pathway. Joint initiative of IITs and IISc – Funded by MHRD Page 69 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Figure 16. Comparison between the SgcE PKS catalyzing the nine-membered enediyne core in C-1027 biosynthesis and the CalD8 PKS catalyzing the 10-membered enediyne core in calicheamicin biosynthesis. aa = amino acid; KS = ketoacyl synthase; AT = acyltransferase; ACP = acyl carrier protein; KR = ketoreductase; DH = dehydratase; and TD = COOH-terminal domain. (Liu, W.; Christenson, S. D.; Standage, S.; Shen, B. Science 2002, 297, 1170.) It is proposed that the SgcE catalyzes the assembly of a nascent linear polyunsaturated intermediate from acetyl and malonyl coenzyme A (CoA) in an iterative process. Next, upon action of other enzymes polyunsaturated intermediate is subsequently desaturated to furnish the two -yne groups and cyclized to afford the enediyne core. An acetylenase has been reported from the plant Crepisalpina that was characterized as a nonheme di-iron protein. However, no such homolog was found within the C-1027 cluster. Close comparison of the C-1027 gene cluster with that of neocarzinostatin gene cluster revealed the presence of a group of ORFs (sgcE1 to sgcE11) in addition to sgcE. These ORFs are highly conserved. The functions of the genes are as follows: (a) Genes SgcE6, SgcE7 and SgcE9 function similar to that of various oxidoreductases. (b) SgcE1, SgcE2, SgcE3, SgcE4, SgcE5, SgcE8 and SgcE11 show either no sequence homology or homology only to proteins of unknown function. (c) SgcE10 is highly homologous to a family of thioesterases. These enzymes, together with the SgcF epoxide hydrolase are responsible for the synthesis of enediyne intermediate 4 from the nascent linear polyunsaturated intermediate (Scheme 7). All other experimental evidences support the polyketide route to the biosynthesis of C-1027 enediyne core. Biosynthesis of Enediyne core of C-1027 O HO O H3C S PCP acetyl CoA + O O SgcE and sgcE1-E11 OH SgcF O O S-CoA malonyl CoA O 4 Enediyne core Scheme 7. Biosynthetic hypothesis for the enediyne core and a convergent assembly strategy. Joint initiative of IITs and IISc – Funded by MHRD Page 70 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Deoxyaminosugar (5) biosynthesis: There are seven genes namely, sgcA, sgcA1, sgcA2, sgcA3, sgcA4, sgcA5, sgcA6 that are responsible for the biosynthesis deoxyaminosugar (5) moiety in C-1027. The seven deoxyaminosugar biosynthesis genes encode a thymine diphosphate (TDP)-glucose synthetase (SgcA1), a TDP-glucose 4,6-dehydratase (SgcA), a TDP4-keto-6-deoxyglucose epimerase (SgcA2), a C-methyl transferase (SgcA3), an amino transferase (SgcA4), an N-methyl transferase (SgcA5) and a glycosyltransferase (SgcA6). Together, they do the enzyme functions that is essential for the biosynthesis of 5 from glucose-1phosphate (Scheme 8) and the attachment of 5 to 4. Biosynthesis of deoxyaminosugar peripheral moiety of C-1027 O OH OH HO HO O SgcA1 HO HO O SgcA O Me HO OH OH OH OP D-Glucose-1-Phosphate OTDP OH OH Me OTDP Me2N O Me OH OH Me O OTDP SgcA4 O SgcA3 O SgcA2 H2N Me OH OH Me O SgcA5 OTDP Me O OH OH Me OTDP 5 deoxyaminosugar moiety Scheme 8. Biosynthetic hypothesis for deoxyaminosugar (5, C). Joint initiative of IITs and IISc – Funded by MHRD OTDP Page 71 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics β-amino acid (6) biosynthesis: There are six genes, sgcC, sgcC1, sgcC2, sgcC3, sgcC4, sgcC5 that encode biosynthesis of β-amino acid (6) in C-1027. The six β-amino acid biosynthesis genes encode (a) a phenol hydroxylase (SgcC), (b) a non ribosomal peptide synthetase (NRPS)– adenylation enzyme (SgcC1), (c) an NRPS peptidyl–carrier protein (PCP) (SgcC2), (d) a halogenase (SgcC3), (e) an aminomutase (SgcC4), and (f) an NRPS-condensation enzyme (SgcC5). These enzyme together functions for the biosynthesis of β-amino acid (6) moiety starting from tyrosine (Scheme 9). Once formed it is activated by SgcC5 as an aminoacyl-S-PCP. The activated aminoacyl-S-PCP is then ready for attachment to the enediyne moiety 4 by SgcC5. Biosynthesis of -amino acid peripheral moiety of C-1027 HO H3N H H H O H H O H H O O sgcC sgcC1 HO HO HO O H3N H H3N H HO sgcC1 O Ad sgcC2 PCP L-Tyrosine NH3 O H H O H H O HO HO S PCP H3N H sgcC3 HO HO S PCP sgcC4 H3N H HO Cl S PCP H HO Cl 6 -amino acid moiety Scheme 9. Biosynthetic hypothesis for β-amino acid (6, D). Joint initiative of IITs and IISc – Funded by MHRD H Page 72 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Benzoxazolinate (7) biosynthesis: Seven genes were identified that are essential for the biosynthesis of benzoxazolinate moiety of C-1027. These are sgcD, sgcD1, sgcD2, sgcD3, sgcD4, sgcD5, sgcD6. These seven benzoxazolinate biosynthesis genes encode the followings: (a) the anthranilate synthase I and II subunits (SgcD and SgcD1), (b) a monoxygenases (SgcD2), (c) a P-450 hydroxylase (SgcD3), (d) an O-methyl transferase (SgcD4), (e) a CoA ligase (SgcD5), and (f) an acyltransferase (SgcD6). Action of these enzymes starts with anthranilate, a common intermediate from the shikimate pathway, for the biosynthesis of benzoxazolinate moiety (7) (Scheme 10). The co-localization of SgcD and SgcD1 gene along with the other C1027 production genes assures the availability of anthranilate for secondary metabolite biosynthesis. However, the origin of the C3 unit is not clear. Moreover, how the C3 unit is fused to the anthranilate intermediate to form the morpholinone unit of benzoxazolinate moiety (7) is also unclear. However, it is believed that the anthranilate unit of benzoxazolinate moiety (7) is activated as an acyl-S-CoA for attachment to the enediyne core 4 by the action of gene SgcD6 (Scheme 11) Biosynthesis of benzoxazolinate peripheral moiety of C-1027 sgcD2 H2N HO HO sgcD3 O OMe "C3 Unit" H2N O OH HO OH O OMe HO H2C O O N H OMe N H O OH H2O CoA O OMe sgcD5 O O OH O H3C O sgcD4 H2N H2N O OH Anthranilic acid OH N H S CoA O OH 7 Benzoxazolinate Scheme 10. Biosynthetic hypothesis for benzoxazolinate (7, E). Joint initiative of IITs and IISc – Funded by MHRD Page 73 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Biosynthesis of C-1027 by attaching the peripheral moieties to the enediyne core: Decoration of Enediyne Core of C-1027 with Peripheral Moieties: Biosynthesis of C-1027 O O O N H OMe CoA O OMe S O N H HO S CoA O SgcD6 Enediyne core 7 Benzoxazolinate SgcD6 Me OH Me O SgcA6 OH OH Me HO Me2N Me2N NH3 O OTDP HO O O S PCP H OH OH SgcA6 OH Me OTDP 5 deoxyaminosugar moiety SgcC5 O H HO Cl 4 Enediyne core MeO O SgcC5 NH3 O HO O S PCP H H HO Cl 6 -amino acid moiety O O N OH OH 1 C-1027 Chromophore O OH O O O Cl Scheme 11. Final step in biosynthesis of C-1027-decoration of enediyne core of C-1027 with peripheral moieties. Joint initiative of IITs and IISc – Funded by MHRD Page 74 of 89 N H O NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.4.5. Biosynthesis of Neocarzinostatin Neocarzinostatin, the first characterized enediyne chromophore, was isolated from the culture filtrate of Streptomyces carzinostaticus var. E-41 as a 1 : 1 complex with an 11 kDa apoprotein. It is the in free-standing state is highly unstable upon exposure to heat, high pH or UV-light irradiation. In comparison to C-1027, neocarzinostatin consists of (a) epoxide-derivatized 9-membered enediyne core — synthesized via polyketide pathway by enediyne PKSs. (b) a distinct deoxy aminosugar — it is derived from glucose-1-phosphate. (c) a naphthoic acid moiety The peripheral moieties of neocarzinostatin are also anchored to the enediyne core but at different positions in comparison to C-1027. This indicates that the two enediyne cores are hydroxylated at different sites during the maturation of the enediyne core (Figure 17). Model of Convergent Biosynthesis of Neocarzinostatin O H3C Napthoic Acid SCoA O ? OMe + O OH SCoA Acetyl-& Malony CoA O O PKSE O O O OH Sodium Bicarbonate O Me O O OH O MeHN HO Me HO HO Me O Enediyne Core O OH O Deoxy Aminosugar H3C OH OP D-Glucose- 1-Phosphate or D-Mannose- 1-Phosphate Glycosyl transferase O PKSE SCoA + OH O SCoA Acetyl-& Malony CoA Figure 17. A model of convergent biosynthesis for the enediyne Neocarzinostatin. Sequencing of the gene cluster ment revealed six complete ORFs (including ncsA) and one incomplete ORF. Remarkably, the four ORFs encode a dNDP-D-mannose synthase (NcsC), dNDP-hexose 4,6-dehydratase (NcsC1, a second distinct NGDH gene in this organism), Nmethyltransferase (NcsC5), and glycosyltransferase (NcsC6). These are the enzymes that would be predicted to be essential for biosynthesis of the deoxy aminosugar moiety of NCS chromophore. Joint initiative of IITs and IISc – Funded by MHRD Page 75 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Enediyne core (4) biosynthesis: Fourteen genes, ncsE to ncsE11 and ncsF1 to ncsF2, were identified within the ncs gene cluster all of which play an important role in the NCS enediyne core biosynthesis. The enediyne core was previously predicted to be synthesized by an iterative type I PKS with five domains, of which the KS, AT, KR, and DH are characteristic of known type I PKSs. NcsE shows head-to-tail sequence homology to the SgcE and CalE8 enediyne PKSs. Consequently, it was proposed that NcsE, in a mechanistic analogy to other enediyne PKSs, catalyzes the formation of the nascent linear polyunsaturated intermediate from one acetyl CoA and seven malonyl CoAs in an iterative manner, which is processed to form the enediyne core by several gene products, including NcsE1–E11 and epoxide hydrolases F1 and F2 (Scheme 12). All reported experimental findings unambiguously established that NcsE is essential for NCS biosynthesis. This further supports an iterative type I PKS paradigm for enediyne core biosynthesis. Biosynthesis of Enediyne core of Neocarzinostatin O H3C S PCP acetyl CoA + O O O ncsE and O O OH ncsF and NcsF2 HO O OH ncsE1-E11 O S-CoA malonyl CoA HO Enediyne core (7x) Scheme 12. Biosynthetic hypothesis for the enediyne core and a convergent assembly strategy to enediyne core of Neocarzinostatin. Naphthoic acid biosynthetic: Naphthoic acid moiety originates from a single polyketide chain of six head-to-tail acetate units. This was revealed from isotopic labeling experiments. Characterisation of neocarzinostatin gene cluster showed that the following enzymes are involved in the biosynthetic pathway for the naphthoic acid moiety: (a) an iterative PKS (NcsB), (b) a CoA ligase and (c) several ancillary enzymes. The biosynthesis of the naphthoic acid moiety starts with NcsB, an iterative PKS that contains the domains like — (a) the ketoacyl synthase (KS), (b) acyltransferase (AT), (c) ketoreductase (KR), (d) dehydratase and (e) acyl carrier protein domain (ACP) and a core domain with unknown function. It is believed that NcsB uses acetyl-CoA as stating matetial and malonyl-CoA as extender to assemble a nascent hexaketide with the selective reduction and dehydration of the keto groups at C5 and C9 (Scheme 13). The hexaketide intermediate then undergoes aromatization to furnish the naphthoic acid via intramolecular aldol condensation. The post-PKS modification of the naphthoic acid moiety starts with the incorporation of a hydroxyl group at C8 carbon which is catalyzed by the cytochrome P450 hydroxylase NcsB3. Ultimately the methylation of the hydroxyl group is catalysed by an S-adenosylmethionine (SAM)-dependent O-methyltransferase (NcsB1). Next, NcsB2 ligase catalyzes the adenylation of 2-hydroxy-7-methoxy-5-methyl-1-naphthoic acid to form its CoA derivative. The discovery of Joint initiative of IITs and IISc – Funded by MHRD Page 76 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics NcsB2 as a promiscuous naphthoic acid/CoA ligase with relaxed substrate specificity provided a great opportunity for producing structural analogs of neocarzinostatin by metabolic engineering. Finally, the putative acyl transferase (NcsB4) is responsible for the transfer of the naphthoic group onto the enediyne core. Scheme 13. Possible biosynthesis mechanism for the naphthoic acid moiety of neocarzinostatin and azinomycin B. Joint initiative of IITs and IISc – Funded by MHRD Page 77 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Deoxyamino Sugar biosynthesis: The deoxyamino sugar moieties of neocarzinostatin and C-1027 differ in the positions and numbers of the amino, hydroxyl and methyl groups. The biosynthetic pathway is likely to start with the activation of the monosaccharide as its NDP derivative by the nucleotidyltransferase NcsC (Scheme 14). Based on sequence homology and established biosynthetic routes, several gene products have been proposed for the activation of the sugar ring via the formation of a 4-keto intermediate, the deoxygenation at C6 and the installation of amino group at C2. Previous labeling experiments with [methyl-3H] methionine have suggested that the N-methyl of the deoxyamino sugar originates from the methionine of Sadenosylmethionine (SAM). The methylation is presumably catalyzed by the methyltransferase NcsC5, whereas the predicted glycosyltransferase NcsC6 may transfer the sugar moiety onto the enediyne core. Together, the characterization of the enzymes in the naphthoic acid and deoxyamino sugar pathways by Shen and coworkers provides an entry point for the exploration of the possibility of producing neocarzinostatin analogs by metabolic engineering and fermentation. The relaxed substrate specificity of NcsB1 and NcsB2 can be potentially exploited. Likewise, the final step catalyzed by the glycosyltransferases NcsC6 could represent another opportunity for generating analogs by glycodiversification. Biosynthesis of deoxyaminosugar peripheral moiety of Neocarzinostatin OH HO HO NcsC (Nucleotidyl transferase) O OH HO HO OH OH OP D-Glucose-1-Phosphate O NcsC1 (Dehydratase) O Me OH Me O HO HO ONDP O NcsC2 O (Dehydratase) ONDP ONDP OH Me NcsC3 (Aminomutase) O Me O OH Me NcsC4 (Dehydratase) HO O HO NH2 O CH3HN ONDP deoxyaminosugar moiety (Methyl transferase) NH2 ONDP NcsC5 HO ONDP Scheme 14. Biosynthetic hypothesis for deoxyaminosugar moiety of neocarzinostatin. Joint initiative of IITs and IISc – Funded by MHRD Page 78 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Final biosynthesis of neocarzinostatin by attaching the peripheral moieties to the enediyne core: A convergent strategy could be envisaged for the assembly of the NCS chromophore from the three individual building blocks of the deoxy-aminosugar, naphthoic acid, and enediyne core (Scheme 15). While the coupling between dNDP-sugar and the enediyne core is catalyzed by the NcsC6 glycosyltransferase, that between naphthoyl-S-NcsB and the enediyne core is most likely catalyzed by the NcsB2 CoA ligase. Although the cyclic carbonyl carbon of NCS has previously been shown to originate from carbonate, no obvious candidate catalyzing the attachment of carbonate could be identified within the gene cluster. The convergent biosynthetic strategy for NCS once again underscores nature‘s efficiency and versatility in synthesizing complex molecules. Scheme 15. Final step in biosynthesis of C-1027-decoration of enediyne core of neocarzinostatin with peripheral moieties. Joint initiative of IITs and IISc – Funded by MHRD Page 79 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.4.6. Biosynthesis of Maduropeptin Maduropeptin was isolated as a 1 : 1 apoprotein–chromophore complex from the Actinomadura madurae strain H710–49 in 1991. The maduropeptin chromophore is tightly bound by a new class of apoprotein that is distinct from the apoproteins of neocarzinostatin, C1027 and kedarcidin. Maduropeptin contains the following units – (a) a 3,6-dimethylsalicyclic acid, (b) a deoxy aminosugar and (c) an aryl hydroxypropionic acid moiety along with the enediyne core. The sequencing and annotation of the maduropeptin gene clusters revealed the presence of biosynthetically important 42 ORFs and the biosynthetic pathways for the three peripheral moieties are related to those of C-1027and neocarzinostatin. Model of Convergent Biosynthesis of Maduropeptin HO HO Me O OH OP D-Glucose- 1-Phosphate H3C SCoA PKS + O O mdpB Deoxy Aminosugar Me HO OH OH Me HN O Enediyne Core O Dimethylsalicyclic acid Glycosyl transferase O OMe Me O O O SCoA Acetyl-& Malony CoA Cl HO O PKSE H3C O SCoA Acetyl-& Malony CoA N O Aryl hydroxypropionic acid H H O H3N H HO O L-Tyrosine Figure 18. A model of convergent biosynthesis for the enediyne, Maduropeptin. Joint initiative of IITs and IISc – Funded by MHRD SCoA + O O Page 80 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Biosynthesis of Dimethylsalicyclic acid peripheral moiety of Maduropeptine: It is known that some bacteria and fungi synthesized 6-methylsalicylic acid (6-MSA) via polyketide synthetic pathway. The presence of a second PKS gene (mdpB) in the maduropeptin gene cluster hinted that the 3,6-dimethylsalicyclic acid moiety may also originate from a polyketide precursor. In addition, MdpB is highly homologous to a group of iterative fungal PKSs that include the 6-methylsalicylic acid synthases (MSAS) from Penicillium patulum, Aspergillus terreus and Glarea lozoyensis. MdpB also shares the same domain composition with NcsB, the PKS for the biosynthesis of the naphthoic acid moiety in neocarzinostatin. The catalytic mechanism involves a partially reduced polyketide intermediate before aromatization (Scheme 16). The selective reduction of the keto group is not clear for these iterative PKSs. The PKS product might undergo methylation and adenylation after it is off loaded from the PKS (pathway a, Scheme 16). In an alternative path, the PKS product may be directly transferred to the sugar moiety with the assistance of a transferase without off-loading (pathway b, Scheme 16). Interestingly, the neighboring gene mdpB3 encodes a hydrolase that may actually act as a transferase. Scheme 16. Possible biosynthetic mechanisms for the 3,6-dimethylsalicyclic acid of maduropeptin. Joint initiative of IITs and IISc – Funded by MHRD Page 81 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Biosynthesis of Aryl hydroxypropionic acid peripheral moiety of Maduropeptine: Study of the maduropeptin gene cluster reveals that the pathway for the (S)-3-(2- chloro-3hdyroxy-4-methoxyphenyl)-3-hydroxypropionic acid or simply the aryl hydroxypropionic acid moiety is resemblance to the biosynthetic pathway for the β-tyrosine moiety of C-1027. The first two enzymes MdpC4 and MdpC1 are highly homologous to the aminomutase SgcC4 and NRPS adenylation domain protein SgcC1. The gene MdpC2 is similar to SgcC2-like PCP to which the substrate is attached (Scheme 17). MdpC7 is a pyridoxal phosphate (PLP)-dependent transaminase that is absent in the biosynthetic pathway of C-1027. It converts the β -amino group into a keto group. Further modification of the aromatic moiety is carried out by the hydrolase MdpC, methyltransferase MdpC6 and halogenase MdpC3. The last enzyme MdpC8 is an alcohol dehydrogenase that furnishes the β-hydroxyl group. In contrast to the ester linkage in C-1027 between the enediyne core and the β-tyrosine moiety, the aryl hydroxypropionic acid moiety is connected to the enediyne core through an amide linkage. The enzyme involved in amide bond formation is not known exactly Biosynthetic pathway for the aryl hydroxypropionic acid moiety of maduropeptin OH OH OH H3N NH3 O O L-Tyrosin O ATP MdpC7 (Transaminase) S O O S OMe HO MdpC6 (Methyl transferase) MdpC (Hydroxylase) NH3 AMP MdpC2 O OH HO OH MdpC1 (NRPS A domain -like protein) MdpC4 (Aminomutase) O HO FAD OMe HO OMe MdpC3 (Halogenase) FADH2 MdpC8 (Dehydrogenase) Cl Cl O FADH2 O S O S O OH FAD O S O S Figure 17. Proposed biosynthetic pathway for the aryl hydroxypropionic acid moiety of maduropeptin. Joint initiative of IITs and IISc – Funded by MHRD O Aryl hydroxypropionic acid moiety Page 82 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Biosynthesis of Aminosugar peripheral moiety of Maduropeptine: Maduropeptin has a different aminosugar compared to C-1027 and neocarzinostatin, with the major difference being the absence of two methyl groups on C5. The synthesis of the aminosugar begins with Dglucose-1-phosphate. The gene product (MdpA1) is highly homologous to the nucleotidyltransferase (SgcA1) and helps installing the NDP group on the D-glucose-1phosphate (Scheme 18). The dehydrogenase (MdpA2) and decarboxylase (MdpA3) are responsible for the removal of the CH2OH group at the C5 position. The sugar nucleotide moiety is further modified by four more enzymes that include (a) a dehydrogenase, (b) a decarboxylase, (c) a SAM-dependent methyltransferase and (d) a transaminase to furnish the final aminosugar. The glycotransferases (SgcA6) finally transfer the sugar moiety onto the enediyne core. Biosynthetic pathway for the Aminosugar moiety of maduropeptin MdpA1 (Nucleotidyltransferase) OH HO HO O OH HO HO HOOC MdpA2 O OH ONDP OP O HO OH OH MdpA3 O (Decarboxylase) O (Dehydrogenase) HO HO OH ONDP ONDP MdpA4 (Methyltransferase) H2N O O MdpA4 (Transaminase) O Me OH Me OH OH ONDP ? OH ONDP Deoxy Aminosugar Dimethyl salicyclic acid Dimethylsalicyclic acid D-Glucose- 1-Phosphate Deoxy Aminosugar Me Me OH HO HN Me O Scheme 18. Proposed biosynthetic pathway for the aminosugar of maduropeptin. Joint initiative of IITs and IISc – Funded by MHRD Page 83 of 89 OH O ONDP NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.5. Biosynthesis of 10-Membered Ring Enediynes Peripheral moieties of the 10-membered enediynes: Based on the structure and folding pattern of the enediyne core the 10-membered enediyne chromophores are further divided into two sub group--(i) calicheamicin-like and (ii) dynemicin-like enediynes. Calicheamicin-like enediynes include (a) esperamicins, (b) namenamicin and (c) shishijimicins. These are decorated with a variety of sugar and aromatic peripheral moieties. In comparison, the dynemicin-like chromophores include (a) dynemicin and (b) uncialamycin. These are characterized by the presence of an anthraquinone peripheral moiety. The calicheamicin and dynemicin gene clusters have been sequenced and partially annotated. Studies revealed the presence of enzymes involved in the biosynthesis of the peripheral moieties of calicheamicin 1I are the sugar-modifying enzymes and glycotransferases. 2.5.1. Biosynthesis of Calicheamicin 1I Calicheamicins were isolated from Micromonospora echinospora spp. calichensis from caliche soils. Calicheamicins represent a subfamily of enediyne chromophores and their structure, mode of action and pharmacological properties have been examined rigorously. Calicheamicin 1I, the best characterized member of the calicheamicins, contains a complex aryltetrasaccharide moiety that confers the chromophore its DNA-binding specificity. The Building blocks of Calicheamicin 1I are (a) enediyne core, (b) aryltetrasaccharide that is composed of an iodized orsellenic acid moiety and four monosaccharides. The aryltetrasaccharide unit is characterized by an unusual thiosugar and a hydroxylamino glycosidic linkage (Figure 19). Model of Convergent Biosynthesis of Calicheamicin 1I SSSMe H3C O SCoA PKSE +O O SCoA Acetyl-& Malony CoA NHCO2Me H HO Glycosyl transferase HO HO O Sugar OH Me O OH N H O Deoxy Aminosugar D O B Sugar O O MeO EtHN OH A Sugar O O Me S O Me OMe I OMe Me O HO MeO OH C Sugar O Orsellenic acid O Enediyne Core O H3C S PCP acetyl CoA NADPH O + O CalO1, CalO6, CalO2, CalO3 Deoxy monosugar O S-CoA malonyl CoA (3x) OH HO HO OH O OH OP OP D-Glucose- 1-Phosphate D-Glucose- 1-Phosphate Figure 19. Building blocks and model of biosynthesis of Calicheamicin 1I. Joint initiative of IITs and IISc – Funded by MHRD Page 84 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Biosynthesis of Orsellenic acid of aryltetrasaccharide peripheral unit of Calicheamicin 1 : In addition to calE8, a second iterative PKS (calO5) gene is present in the Calicheamicin 1I cluster. The predicted gene product (CalO5) shares significant sequence homology with AviM, the iterative PKS for the biosynthesis of the oligosaccharide antibiotics avilamycins produced by Streptomyces viridochromogenes. Notably, AviM is also the first type I polyketide synthase isolated from bacteria to produce an aromatic compound. The fact that avilamycins and the related evernimicin also contain a chlorinated orsellenic acid group hints that CalO5 could be involved in the synthesis of the iodized orsellenic moiety in calicheamicin. I Sequence analysis suggested that CalO5 contains at least four domains (KS, AT, DH and ACP) with a ‗core‘ domain apparently located between the DH and ACP domains. The lack of KR domain suggests that the keto groups remain unreduced in the nascent polyketide chain. A possible mechanism for the formation of the aromatic ring has been suggested (Scheme 19). Four predicted enzymes that include a hydroxylase, a halogenase and two methyltransferases putatively modify the aromatic ring to furnish the final product. The predicted cytochrome P450 (CalO2) and FADH2-dependent halogenase (CalO3) are likely to catalyze the hydroxylation and iodination respectively, whereas the O-methyltransferases CalO1 and CalO6 install the two methyl groups. Although the precise timing of the post-PKS steps and the stringency of the substrate specificity of the enzymes remain to be fully established, biochemical and structural characterization of CalO2 indicated that the hydroxylation may occur after the halogenations. Based on the crystal structure of CalO2, MoCoy and coworkers also suggested that CalO2 may act on a CoA-derivatized or ACP domain-tethered substrate. Thorson and coworkers have also confirmed that sugar C is installed on the orsellenic group by glycosyltransferase CalG1. Meanwhile, the FabH/KS domainlike CalO4 is likely to catalyze the formation of the thioester linkage between the orsellenic acid and sugar B. Scheme 19. Putative biosynthetic mechanism for the orsellenic acid moiety of calicheamicins. Joint initiative of IITs and IISc – Funded by MHRD Page 85 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Biosynthesis of the sugar moieties of calicheamicin 1I: Biosynthesis of monosaccharides-The Aryl Tetrasaccharide of Calicheamicin 1I: Calichiamicin 1I contains four highly modified monosaccharides. Two of the monosaccharides are linked via an unusual hydroxylamino glycosidic linkage. All four sugars are deoxysugars that exhibit greater hydrophobicity compared to common sugars. The deoxysugars along with the linked orsellenic acid group is giving the DNA binding specificity of calicheamicin 1I. The biosynthesis of the four monosaccharides starts with the conversion of glucose-1-phosphate to NDP-glucose which is similar to the biosynthetic pathway of sugar for C-1027, neocarzinostatin, maduropeptin and other glycosides. Analysis of gene cluster of Calichiamicin 1I reveals that CalS7 catalyzes the formation of NDP-glucose in the early stage of synthesis of sugars A–D. As discussed below, the pathways for the four sugars would diverge after the formation of NDP-glucose. The 4hydroxyamino-6-deoxy--D-glucose (sugar A) is directly tethered to the enediyne core. The deoxygenation at C6 position and installation of the amino group are followed similar routes that involve the NDP-4-keto-6-deoxy--glucose intermediate in case of C-1027 (Scheme 20). Biosynthesis of Monosugars peripheral moietes of Calichiamicin AT2433-B1 D-Glucose- 1Phosphate CalS7 CalS8 CalS3 NDP-4-keto-6deoxy--D-glucose CalS9 CalS14 CalE10 CalS12/13 Sugar A CalS10 Sugar B Sugar C Scheme 20. Biosynthetic pathways for the monosaccharides of calicheamicin 1I (Liang, Z-X Nat. Prod. Rep. 2010, 27, 499.). Joint initiative of IITs and IISc – Funded by MHRD Page 86 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics The hydroxylamino glycosidic bond between sugar A and sugar B is very crucial for maintaining the correct conformation and rigidity of the oligosaccharide for effective binding to the minor groove of DNA. A novel aminosugar oxidase, the N-oxidase CalE10, enzyme catalyzes the hydroxylation of the amine group. CalE10 is a heme protein that exhibits stringent regiospecificity with limited overoxidation in vitro. Calicheamicin 1I contains a 4-thio-2,4,6-trideoxy-D-glucose (sugar B) that is connected to the orsellenic acid moiety through a thioester linkage. The sulfur atom of sugar B contributes to the binding mode of calicheamicin 1I by forming hydrogen bonds with an exposed amino proton of guanine in the minor groove of DNA. However the mechanism of installation of sulfur atom is not known. The closely related esperamicin A1 also contains a similar thiosugar. From the isotope feeding experiment, it is known that the sulfur is derived from L-methionine or Lcysteine. The enzymes involved in the biosynthesis of methylated 3-methoxy-L-rhamnose (sugar C) synthesis remains unclear. However, it is believed that the biosynthesis follow a common pathway for rhamnose synthesis with the deoxygenation at C6 facilitated by the formation of the NDP-4-keto-6-deoxy--D-glucose intermediate (Scheme 20). A putative epimerase and a methyltransferase most probably catalyze the epimerization at C2 and methylation at C3 respectively. Aminodideoxypentose (4-amino-3-O-methoxy-2,4,5-trideoxypentose, sugar D) present in calicheamicin 1I is similar to that present in esperamicin A1 and in indolocarbazole antitumor antibiotic AT2433. Comparison of the calicheamicin and AT2433 gene clusters indicated the presence of a set of common genes that are expectedly involved in the biosynthesis of the 4amino-3-O-methoxy-2,4,5-trideoxypentose. It is established that the synthesis of the modified pentose proceeds via a TDP-sugar pathway. A decarboxylation step is involved at C6 position with the help of a UDP-glucuronate decarboxylase-like protein (CalS9). The deoxygenation of C2 and installation of amino group at C4 follow as per the mechanisms shown in Scheme 20. Assembly of calicheamicin 1I by glycosyltransferases: The orsellenic acid and sugar moieties are assembled onto the enediyne aglycone in the final stages of calicheamicin biosynthesis. A set of O-glycosyltransferases are utilized to catalyze the transfer of the sugars from the nucleotide diphosphate sugar donors to the aglycone acceptor. Glycosyltransferases CalG3 and CalG2 catalyze the first two glycosylation steps in a sequential fashion (Figure 21). The crystal structure of CalG3 revealed a typical UDP-glycosyltransferase/glycogen phosphorylase fold found in many glycosyltransferases. It is also known that CalG3 catalyzes the formation of the unusual hydroxylamino glycosidic bond by using TDP-4,6-dideoxy-a-D-glucose as a substrate surrogate. The other two putative glycosyltransferases (CalG1 and CalG4) encoded by the gene clusters are the rhamnosyltransferase and aminopentosyltransferase which are responsible for the installation of sugars C and D respectively. Joint initiative of IITs and IISc – Funded by MHRD Page 87 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics Figure 21. Assembly of calicheamicin 1I by glycosyltransferases (Liang, Z-X Nat. Prod. Rep., 2010, 27, 499.). Joint initiative of IITs and IISc – Funded by MHRD Page 88 of 89 NPTEL – Chemistry – Bio-Organic Chemistry of Natural Enediyne Anticancer Antibiotics 2.6. Selected References 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. Smith, A. L.; Nicolaou, K. C. J. Med. Chem. 1996, 39, 2103‐2117. (a) Nicolaou, K. C.; Dai, W. M. Angew. Chem., Int. Ed. Engl. 1991, 30, 1034. (b) Myers, A. G.; Proteau, P. J. J. Am. Chem. Soc. 1989, 111, 1147‐1148. Myers, A. G.; Tom, N. J.; Fraley, M. E.; Cohen, S. B.; Madar, D. J. J. Am. Chem. Soc. 1997, 119, 6072‐6094 18. Shair, M. D.; Yoon, T. Y.; Mosny, K. K.; Chou, T. C.; Danishefsky, S. J. J. Am. Chem. Soc. 1996, 118, 9509‐9525. Taunton, J.; Wood, J. L.; Schreiber, S. L. J. Am. Chem. Soc. 1993, 115, 10378‐10379. Kobayashi, S.; Ashizawa, S.; Takahashi, Y.; Sugiura, Y.; Nagaoka, M.; Lear, M. J.; Hirama, M. J. Am. Chem. Soc. 2001, 123, 11294. Ji, N.; O‘Dowd, H.; Rosen, B. M.; Myers, A.G. J. Am. Chem. Soc. 2006, 128, 14825‐14827. Ji, N.; O‘Dowd, H.; Rosen, B. M.; Myers, A.G. J. Am. Chem. Soc. 2006, 128, 14825‐14827. Dai, W.‐M.; Fong, K. C.; Lau, C. W.; Zhau, L.; Hamaguchi, W.; Nishimoto, S.‐i. J. Org. Chem. 1999, 64, 682. Basak, A.; Roy, S. K.; Mandal, S. Angew. Chem., Int. Ed. 2005, 44, 132. Basak, A.; Kar, M.; Mandal, S. Bioorg. Med. Chem. Lett. 2005, 15, 2061. Zhao, Z.; Peacock, J. G.; Gubler, D. A.; Peterson, M. A. Tetrahedron Lett. 2005, 46, 1373. Nicolaou, K. C.; Dai, W.‐M. J. Am. Chem. Soc. 1992, 114, 8908. Poloukhtine, A.; Popik, V. V. Chem. Commun. 2005, 617. Kar, M.; Basak, A.; Bhattacharya, M. Bioorg. Med. Chem. Lett. 2005, 15, 5392. (a) Lanen, S. G. V.; Shen, B. Curr. Top. Med. Chem. 2008, 8, 448–459 and references therein. (b) Liu, W.; Christenson, S. D.; Standage, S.; Shen, B. Science 2002, 297, 1170. (c) Liang, Z.-X. Nat. Prod. Rep. 2010, 27, 499–528 and references therein. Joint initiative of IITs and IISc – Funded by MHRD Page 89 of 89