Slides - BITS
advertisement

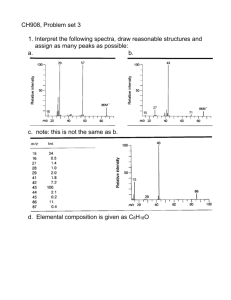
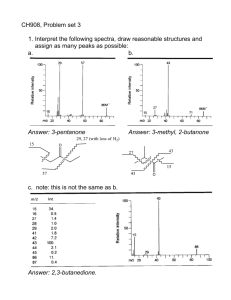
introduction to mass spectrometry and proteomics lennart martens lennart.martens@vib-ugent.be computational omics and systems biology group VIB / Ghent University, Ghent, Belgium Advertorial Amino Acids and Proteins Mass Spectrometry: Concepts and Components Tandem Mass Spectrometry (MS/MS, MS²) A CID Fragmentation Primer Mass Spectrometer Configurations Proteomics Methods: 2D PAGE Shotgun (Gel-Free) Approaches Workhorse Shotgun Method: MudPIT Amino Acids and Proteins Mass Spectrometry: Concepts and Components Tandem Mass Spectrometry (MS/MS, MS²) A CID Fragmentation Primer Mass Spectrometer Configurations Proteomics Methods: 2D PAGE Shotgun (Gel-Free) Approaches Workhorse Shotgun Method: MudPIT Amino Acids and their properties From: http://courses.cm.utexas.edu/jrobertus/ch339k/overheads-1/ch5-amino-acids.jpg A protein backbone H H O side chain R1 H C N H C + H C C N O H H H O N H2N H O R2 H O residue C N C O H C C O R2 H N O R7 O H R5 N R4 O peptide bond O N N H H H R3 O H R1 N carboxyl group R1 amino terminus O O amino group + R2 N R6 H O OH carboxyl terminus A protein sequence N H2N O N N R2 O H N N R4 O H R7 O H R5 O H R3 O H R1 O N R6 OH H Methionine Glycine Alanine Serine Tyrosine Leucine Arginine Met Gly Ala Ser Tyr Leu Arg M G A S Y L R MGASYLR Amino Acids and Proteins Mass Spectrometry: Concepts and Components Tandem Mass Spectrometry (MS/MS, MS²) A CID Fragmentation Primer Mass Spectrometer Configurations Proteomics Methods: 2D PAGE Shotgun (Gel-Free) Approaches Workhorse Shotgun Method: MudPIT Schematic view of a generalized mass spec sam ple ion source m ass analyzer(s) detector digitizer Generalized mass spectrometer All mass analyzers operate on gas-phase ions using electromagnetic fields. Results are therefore plotted on a cartesian system with mass-over-charge (m/z) on the X-axis and ion intensity on the Y-axis. The latter can be in absolute or relative measurements. The ion source therefore makes sure that (part of) the sample molecules are ionized and brought into the gas phase. The detector is responsible for actually recording the presence of ions. Time-of-flight analyzers also require a digitizer (ADC). Ion sources: MALDI h ⋅ν laser irradiation desorption analyte target surface matrix molecule high vacuum + + + + + + + + + + H+ proton transfer + + + + + + + + + + + + + + + + Gas phase Matrix Assisted Laser Desorption and Ionization (MALDI) MALDI sources for proteomics typically rely on a pulsed nitrogen UV laser (υ = 337 nm) and produce singly charged peptide ions. Competitive ionisation occurs. The term ‘MALDI’ was coined by Karas and Hillenkamp (Anal. Chem., 1985) and Koichi Tanaka received the 2002 Nobel Prize in Chemistry for demonstrating MALDI ionization of biological macromolecules (Rapid Commun. Mass Spectrom., 1988) MALDI matrix molecules Matrix properties: • Small organic molecules • Co-crystallize with the analyte • Need to be soluble in solvents compatible with analyte • Absorb the laser wavelength • Cause co-desorption of the analyte upon laser irradiation • Promote analyte ionization MALDI – spotting on target + matrix sample matrix + sample 1-2 mm spot on target and let air-dry stainless steel, gold, teflon, … matrix crystals Ion sources: ESI m/z analyzer inlet www.sitemaker.umich.edu/mass-spectrometry/sample_preparation + + + 3-5 kV + + + + + + + + + + + + + + + + droplet evaporation and charge-driven fission or ion expulsion + + sam ple N2 N2 + 0 needle nebulisation Electospray ionization (ESI) + 0 0 evaporation only + 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 barrier ESI sources typically heat the needle to 40°to 100°to facilitate nebulisation and evaporation, and typically produce multiply charged peptide ions (2+, 3+, 4+) John B. Fenn received the 2002 Nobel Prize in Chemistry for demonstrating ESI ionization of biological macromolecules (Science, 1989) – ESI is also used in fine control thrusters on satellites and interstellar probes… ESI – online LC and solvents (nano) RP column solvent mixer aqueous solvent H2O + 0.1% FA + 2–5% ACN A B (100–5 µm) (nano) needle spray organic solvent ACN + 0.1% FA Nanospray ESI sources (5-10 µm diameter needle) achieve a higher sensitivity, probably due to the higher surface-to-volume ratio. For a spherical droplet this ratio is: A = 4 ⋅π ⋅ r V =4 2 A 3 6 = = V r D π ⋅ r3 3 Analyzers: time-of-flight (TOF) high vacuum sample ions source extraction plate (30 kV) Ek = q ⋅ V detector field-free tube (time-of-flight tube) > 1 meter 2 ⋅ Ek m ⋅ v2 ↔v= Ek = 2 m We can now relate m/q (or the more commonly used m/z) to the velocity of the ion, and using Newton’s kinematica we can relate the speed to the travel time and (known + exactly calibrated) field-free tube length m 2 ⋅V 2 ⋅V 2 ⋅V ⋅ t 2 = 2 = = 2 q v x2 x t Analyzers: time-of-flight – reflectron Ekin > Ekin ion mirror reflectron detector with reflectron Analyzers: time-of-flight: delayed extraction source extraction plate (0 kV) source extraction plate (0 kV) field gradient **** *** ** * source extraction plate (30 kV) time t0 tx t x + 100 ns Resolution and why it matters Resolution in mass spectrometry is usually defined as the width of a peak at a given height (there is an alternative definition based on percent valley height). This width can be recorded at different heights, but is most often recorded at 50% peak height (FWHM). monoisotopic mass average mass From: Eidhammer, Flikka, Martens, Mikalsen – Wiley 2007 Analyzers: ion trap (IT) DC/ACRF voltage source detector capping electrode ring electrode capping electrode Ion traps operate by effectively trapping the ions in an oscillating electrical field. Mass separation is achieved by tuning the oscillating fields to eject only ions of a specific mass. Big advantages are the ‘archiving’ during the analysis, allowing MSn. Wolfgang Paul and Hans Georg Dehmelt received the 1989 Nobel Prize in Physics for the development of the ion trap. Analyzers: quadrupole (Q) + (U + V ⋅ cos ωt ) permitted m/z ejected m/z − (U + V ⋅ cos ωt ) ejected m/z Quadrupole mass analyzers also use a combined RF AC and DC current. They thus create a high-pass mass filter between the first two rods, and a low-pass mass filter between the other two rods. The net result is a filter that can be fine-tuned to overlap (and thus permit) only in a specific m/z interval; ions of all other m/z values will be ejected. Detectors: electron multiplier single ion in 40V 20V 80V 60V 120V 100V 10 6 electrons out Different variations of electron multiplier (EM) detectors are in use, and they are the most common type of detector. An EM relies on several Faraday cup dynodes with increasing charges to produce an electron cascade based on a few incident ions. Fourier transform ion cyclotron resonance (FTICR) electrodes ion orbit strong magnetic field An FT-ICR is essentially a cyclotron, a type of particle accelerator in which electrons are captured in orbits by a very strong magnetic field, while being accelerated by an applied voltage. The cyclotron frequency is then related to the m/z. Since many ions are detected simultaneously, a complex superposition of sine waves is obtained. A Fourier transformation is therefore required to tease out the individual ion frequencies. Orbitrap From: http://www.univ-lille1.fr/master-proteomique/proteowiki/index.php/Orbitrappe An OrbiTrap is a special type of trap that consists of an outer and inner coaxial electrode, which generate an electrostatic field in which the ions form an orbitally harmonic oscillation along the axis of the field. The frequency of the oscillation is inversely proportional to the m/z, and can again be calculated by Fourier transform. The OrbiTrap delivers near-FT-ICR performance, but is cheaper, much more robust, and much simpler in maintenance. It is a recent design, only a few years old. Amino Acids and Proteins Mass Spectrometry: Concepts and Components Tandem Mass Spectrometry (MS/MS, MS²) A CID Fragmentation Primer Mass Spectrometer Configurations Proteomics Methods: 2D PAGE Shotgun (Gel-Free) Approaches Workhorse Shotgun Method: MudPIT Tandem-MS: the concept source detector ion selector fragm ent m ass analyzer fragm entation Tandem-MS is accomplished by using two mass analyzers in series (tandem) (note that a single ion trap can also perform tandem-MS). The first mass analyzer performs the function of ion selector, by selectively allowing only ions of a given m/z to pass through. The second mass analyzer is situated after fragmentation is triggered (see next slides) and is used in its normal capacity as a mass analyzer for the fragments. Why tandem-MS? peptide structure x3 y3 R1 NH2 C H a1 CO b1 N H c1 z3 x2 y2 z2 R2 R3 CH2 CH2 C H CO a2 b2 N H c2 C H x1 z1 R4 CO a3 y1 b3 N H C H COOH c3 There are several other ion types that can be annotated, as well as ‘internal fragments’. The latter are fragments that no longer contain an intact terminus. These are harder to use for ‘ladder sequencing’, but can still be interpreted. This nomenclature was coined by Roepstorff and Fohlmann (Biomed. Mass Spec., 1984) and Klaus Biemann (Biomed. Environ. Mass Spec., 1988) and is commonly referred to as ‘Biemann nomenclature’. Note the link with the Roman alphabet. Creating fragments (unimolecular) laser (( )) activated ion (metastable) non-activated ion (stable) precursor ion fragment ions 200 400 600 800 1000 1200 1400 1600 m/z This fragmentation method is called post-source decay (PSD) and relies on a single unimolecular event, in which a highly energetic (metastable) ion spontaneously fragments. PSD typically causes backbone fragmentation. y and b ions are by far the most prevalent fragment types, although TOF-TOF instruments (see later) specifically also yield substantial internal fragments. Creating fragments (bimolecular I) gas inlet collision cell collision gas (atom or molecule) selected peptide ∆V This fragmentation method is called collision-induced dissociation (CID) and relies on a series of bimolecular events (collisions) to provide the peptide precursor with sufficient energy to fragment. CID typically causes backbone fragmentation. y and b ions are by far the most prevalent fragment types. The collision gas is typically an inert noble gas (e.g.: Ar, He, Xe), or N2. Creating fragments (bimolecular II) electron source fragm entation cell - - - - - electron - selected peptide ∆V This fragmentation method is called electron-capture dissociation (ECD) or electrontransfer dissociation (ETD) and relies on a single impact of an electron on a peptide precursor. This high-speed impact immediately imparts sufficient energy to fragment the precursor (non-ergodic process). Like CID, ETD and ECD typically cause backbone fragmentation, but they typically result in c and z ions. ECD is only workable in FT-ICR mass spectrometers, whereas ETD is used in traps. Amino Acids and Proteins Mass Spectrometry: Concepts and Components Tandem Mass Spectrometry (MS/MS, MS²) A CID Fragmentation Primer Mass Spectrometer Configurations Proteomics Methods: 2D PAGE Shotgun (Gel-Free) Approaches Workhorse Shotgun Method: MudPIT Al an i Ar n e gi As ni ne p As ara pa gin rti e c A C cid y G lu ste ta m ine ic G Ac i lu d ta m in G e ly c H ine is ti Is din ol e eu ci n Le e uc in e Ly si M n e Ph thio e en ni yl ne al an in Pr e ol in e Se Th rin re e Tr oni yp ne to ph Ty an ro si ne Va lin e CID fragmentation: the mobile proton model Assum ption peptide Hypothesis peptide pK a values 12 10 pKa 4 2 NH2– H+ X H+ fragmentation event 14 R–group 8 6 –COOH 0 Illustration of the MPM hypothesis The necessary collision energy to achieve a given fragmentation efficiency is highest and equal for both 1+ peptides on the left (the R present in both sequesters the charge), and is higher for PPGFSPFR for the 2+ peptides. This is because the latter has an N-terminal P, which has a relatively high pKa for the N-terminus; so the second available charge is located at either end of the peptide. The other peptide has no real main contender (in pKa) for the second charge, and therefore fragments more easily. On the right, having two R’s in the sequence negates the effect of double charge (both charges are sequestered by an R). From: Wysocki et al, J. Mass Spectrom., 2000 Ionization and fragmentation 1+ peptide fragmentation b-ion y-ion 1+ ? 2+ peptide fragmentation 1+ b-ion y-ion 1+ Amino Acids and Proteins Mass Spectrometry: Concepts and Components Tandem Mass Spectrometry (MS/MS, MS²) A CID Fragmentation Primer Mass Spectrometer Configurations Proteomics Methods: 2D PAGE Shotgun (Gel-Free) Approaches Workhorse Shotgun Method: MudPIT ESI ion trap ESI source ion trap detector Very simple and reliable instrument, that can perform MS and CID MS/MS thanks to the ion trap. Mass accuracy is relatively poor however, and the resolution is lacking as well (unable to distinguish isotopes, hampering charge state determination). ESI triple quadrupole ESI source quadrupole 1 quadrupole 2 quadrupole 3 detector Simple instrument, that recently attracted attention because it is well-suited for Multiple Reaction Monitoring (MRM). It can perform MS and MS/MS, where the first quadrupole is the ion selector, the second quadrupole a collision cell and the third quadrupole a mass analyzer. Due to the ‘wastefulness’ of a quadrupole as mass analyzer, it is not very popular for general MS/MS analysis, however. MALDI DE RE-TOF reflectron detector M ALDI source delayed ex traction TOF tube linear detector reflectron voltage gate for precursor selection Relatively simple instrument, that can measure intact masses quite accurately and can perform fragmentation analysis through PSD (unimolecular fragmentation), but the yield of fragmentation is poor. Cheap and reliable, but out of date nowadays. MALDI TOF-TOF reflectron detector M ALDI source delayed ex traction linear detector source 2 TOF 1 TOF 2 reflectron voltage gate for precursor selection A modern version of the MALDI DE RE-TOF, the TOF-TOF relies on two TOF tubes in tandem. The second TOF is fitted with a reflectron. Mass accuracy and resolution are very high and the instrument can perform MS and MS/MS, both for peptides as well as whole proteins. The archiving nature of the MALDI targets allows the instruments to scan a single sample more thoroughly. TOF-TOF instruments are also well-suited for profiling. ESI quadrupole time-of-flight (Q-TOF) pusher ESI source quadrupole collision cell detector TOF reflectron One of the first hybrid mass spectrometers, the Q-TOF combines the excellent precursor selection characteristics of the quadrupole with the mass resolution and accuracy of the TOF tube. The ESI source produces multiply charged ions, and can be coupled to an online (nano-) LC separation. A Q-TOF is a very good instrument when high-quality data are more important than high-throughput, and is well suited for protein characterisation. ESI quadrupole linear ion trap source detector capping quadrupole capping electrode electrode Linear ion traps store ions in a fixed linear trajectory rather than in a ‘blob’. The major advantage is the increased capacity for trapping ions. This allows for a broader dynamic range and better quantitation performance. The linear ion trap is these days often encountered as the workhorse mass spectrometer in many labs, and can be extended with the highly accurate orbitrap or FT-ICR mass analyzers/detectors. ESI linear ion trap FT-ICR or Orbitrap C-trap ESI source linear ion trap FT-I CR Orbitrap The combination of a linear ion trap and a high-resolution, high-accuracy FT analyzer allows for a broad dynamic range and highly accurate mass measurements. Since the ion trap can be used as a collision cell, the FT analyzer can also measure the resulting fragments with high accuracy. Amino Acids and Proteins Mass Spectrometry: Concepts and Components Tandem Mass Spectrometry (MS/MS, MS²) A CID Fragmentation Primer Mass Spectrometer Configurations Proteomics Methods: 2D PAGE Shotgun (Gel-Free) Approaches Workhorse Shotgun Method: MudPIT 2D-PAGE separation of proteins (Est. 1975) P rinciple cell lysis protein ex traction cells Protein A Protein C Protein B Protein D protein m ix ture pI Chem istry toolbox Mr 2D-PAGE 2D-PAGE separation of proteins (Est. 1975) protein complex protein mixture extraction http://www.akh-wien.ac.at/biomed-research/htx/platweb1.htm 2D-PAGE separation MS/MS analysis 100 pI % 0 100 300 500 700 900 1100 1300 1500 1700 1900 2100 m/z fragmentation 100 MS analysis tryptic % 0 digest 300 400 500 600 700 800 900 1000 1100 m/z MW Amino Acids and Proteins Mass Spectrometry: Concepts and Components Tandem Mass Spectrometry (MS/MS, MS²) A CID Fragmentation Primer Mass Spectrometer Configurations Proteomics Methods: 2D PAGE Shotgun (Gel-Free) Approaches Workhorse Shotgun Method: MudPIT Overall gel-free proteomics workflow protein extraction complex protein mixture http://www.akh-wien.ac.at/biomed-research/htx/platweb1.htm Data-dependent M S/ M S analyses 100 100 100 % % % 0 100 300 500 700 900 1100 1300 1500 1700 1900 2100 m/z 0 100 300 500 700 900 1100 1300 1500 1700 1900 2100 m/z 0 100 300 500 700 900 1100 1300 1500 1700 enzymatic digest extremely complex peptide mixture 1900 2100 m/z MS analysis separation selection less complex peptide fractions Going gel-free in the new millennium • ICAT (Gygi et al., 1999) • MudPIT (Washburn et al., 2001) • Accurate Mass Tags for proteome analysis (Conrads et al., 2000) • Signature Peptides approach for proteomics (Ji et al., 2000) • AA-based covalent chromatography peptide selection (Wang & Regnier, 2001) • Affinity-based enrichment of phosphopeptides (Oda et al., 2001) • ICAT for phosphopeptides (Zhou et al., 2001) • Reversible biotinylation of Cys-peptides (Spahr et al., 2000) • COFRADIC (Gevaert et al., 2002) Amino Acids and Proteins Mass Spectrometry: Concepts and Components Tandem Mass Spectrometry (MS/MS, MS²) A CID Fragmentation Primer Mass Spectrometer Configurations Proteomics Methods: 2D PAGE Shotgun (Gel-Free) Approaches Workhorse Shotgun Method: MudPIT MudPIT: that which we call a rose… Strong cation ex changer SCX R everse-phase resin RP ESI-based MS • Orthogonal, 2D separation of peptides • 2D analogon: pI = SCX, Mr = RP But what about the complexity? e.g., Escherichia coli 4,349 predicted proteins if 100% expressed 109,934 detectable tryptic peptides if 50% expressed 54,967 detectable tryptic peptides Sample complexity increased one order of magnitude! Thank you! Questions?