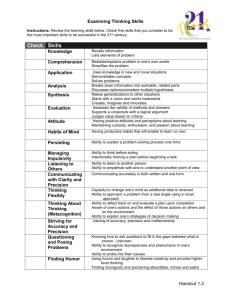
# In R the pound sign denotes a comment line. > # I choose to

> # In R the pound sign denotes a comment line.
> # I choose to include extra spaces for legibility, e.g. my use of the assign command <- in some examples below.
> # If you hit ‘return’ intending to ‘change line’ R will abruptly execute as it (R just made its own line change) will when I hit return after the next word
>
> # Many R users like to prepare code in a text processor then execute parts or all of it by dropping that into R and hitting return. After the next prompt I will paste in the following and hit return. This is really useful. You will see the original paste in until you hit return whence it will be reformatted for display of the results.
pi
pi^2
> pi
[1] 3.141593
> pi^2
[1] 9.869604
> # Here is a snapshot of how it looks in R after paste before you hit return.
> # R knows some physical constants which it prints to default or specified accuracy.
> pi
[1] 3.141593
> print(pi^2, digits = 9)
[1] 9.8696044
> # Function w, defined just below, will print pi to a specified number of digits n, defaulting to n = 10 if n is not specified.
> w <- function(n=10) {print(pi, digits = n)}
> w function(n=10) {print(pi, digits = n)}
> w()
[1] 3.141592654
> w(22)
[1] 3.141592653589793115998
> w(23)
Error in print.default(pi, digits = n) : invalid ‘digits’ argument
> # R has simplified notation for much needed constructs.
> x <- 1:100
> x
[1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14
[15] 15 16 17 18 19 20 21 22 23 24 25 26 27 28
[29] 29 30 31 32 33 34 35 36 37 38 39 40 41 42
[43] 43 44 45 46 47 48 49 50 51 52 53 54 55 56
[57] 57 58 59 60 61 62 63 64 65 66 67 68 69 70
[71] 71 72 73 74 75 76 77 78 79 80 81 82 83 84
[85] 85 86 87 88 89 90 91 92 93 94 95 96 97 98
[99] 99 100
> length(1:10000000)
[1] 10000000
> # Data input by hand is easily done as follows (you respond to the prompts [1], ..etc).
Remember, R does the carriage returns. You use return to execute. I used ‘return’ when prompted with [13]: below. I could have done so after entering the value 3.
>v <- scan()
1: 3 5 8 0 -2 4.4
7: 9.9 11 8 0
11: 0 3
13:
Read 12 items
> v
[1] 3.0 5.0 8.0 0.0 -2.0 4.4 9.9 11.0 8.0 0.0 0.0 3.0
> # You can query R about functions using ? or features using ??.
> # Query ‘?mean’ is recognized by R and returns information about the mean function in R.
I’ve copied part of the output to a comment below. It reveals that ‘mean’ is really the trimmed mean and it can deal with missing data, defaulting to mean when not called with additional arguments.
> ? mean
> # Usage mean(x, ...)
## Default S3 method: mean(x, trim = 0, na.rm = FALSE, ...)
> mean(x)
[1] 50.5
> mean(1:100)
[1] 50.5
> mean(v)
[1] 4.191667
> mean(x^pi)
[1] 473113.7
>y <- c(88, 1:10)
> y
[1] 88 1 2 3 4 5 6 7 8 9 10
> mean(y)
[1] 13
>c(mean(y), mean(y, trim= 0.1))
[1] 13 6
> # Query ‘?? precision’ will be recognized by the system and returns a lot of information about precision in R. I’ve copied part of the output to a comment below.
> ??precision
> # All R platforms are required to work with values conforming to the IEC 60559 (also known as IEEE 754) standard.
This basically works with a precision of 53 bits, and represents to that precision a range of absolute values from about
2e-308 to 2e+308. It also has special values NaN (many of them), plus and minus infinity and plus and minus zero
(although R acts as if these are the same). There are also denormal(ized) (or subnormal) numbers with absolute values above or below the range given above but represented to less precision.
> # Query ‘?precision’ is not recognized by R since precision is not a function in R.
> ?precision
No documentation for ‘precision’ in specified packages and libraries:you could try
‘??precision’
> xdat <- c(1,2.3,5.1,1,3.0,4.7,1,1.6,3.5,1,2.9,3.7)
> xdat
[1] 1.0 2.3 5.1 1.0 3.0 4.7 1.0 1.6 3.5 1.0 2.9 3.7
> xmat <- matrix(xdat,4,3,T) # T is for row-wise scan
> xmat
[,1] [,2] [,3]
[1,] 1 2.3 5.1
[2,] 1 3.0 4.7
[3,] 1 1.6 3.5
[4,] 1 2.9 3.7
> xmat[1,] # extract row 1
[1] 1.0 2.3 5.1
> xmat[1,2] # extract row 1 column 2 entry
[1] 2.3
> xmat[,2] # extract column 2
[1] 2.3 3.0 1.6 2.9
> xmat
[,1] [,2] [,3]
[1,] 1 2.3 5.1
[2,] 1 3.0 4.7
[3,] 1 1.6 3.5
[4,] 1 2.9 3.7
> xmat[1,2]
[1] 2.3
> xmat[1,]
[1] 1.0 2.3 5.1
> xmat[,2]
[1] 2.3 3.0 1.6 2.9
> # ‘?? transpose’ tells us that function ‘t’ transposes a matrix.
> # dot product is %*%
> xmat[1,]%*%xmat[2,] # dot product of rows 1 and two
[,1]
[1,] 31.87
> xmat%*%t(xmat) # dot products of all pairs of rows
[,1] [,2] [,3] [,4]
[1,] 32.30 31.87 22.53 26.54
[2,] 31.87 32.09 22.25 27.09
[3,] 22.53 22.25 15.81 18.59
[4,] 26.54 27.09 18.59 23.10
> # ordinary linear regression of ydat on design matrix xmat is simply
betahat <- ginv(xmat)%*%ydat (betahat is the vector of coeff of LS fit) where ginv is the Moore-Penrose generalized inverse of xmat. Don’t be concerned if you’ve not studied it. Just consider it a ‘black box’ op for now.
(Maybe consider it a miracle to have at hand for free!)
> # the ‘fitted values of regression’ are given by
xmat%*%betahat
> # Query ‘?ginv’ tells me to query ‘??ginv’ which says to load a package MASS which has ginv.
> require(MASS)
> betahat <- ginv(xmat)%*%ydat
> betahat
[,1]
[1,] 14.2085921
[2,] -0.3985031
[3,] -1.5428846
> yfitted <- xmat%*%betahat
> yfitted
[,1]
[1,] 5.423324
[2,] 5.761525
[3,] 8.170891
[4,] 7.344260
> # I’ve inserted the plot of pairs (yfitted, ydat) as a graphic here. In R it will be shown outside the command window. For a perfect fit the points will be on the line ydat = yfitted.