Enzymes: Catalytic Strategies
advertisement

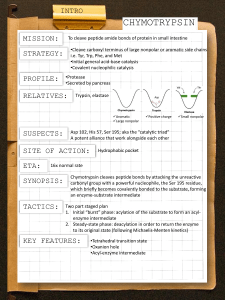
BIOC 460 Summer 2010 Enzymes: Catalytic Strategies Reading: Berg, Tymoczko & Stryer, 6th ed., Chapter 9, pp. 241-254 hexokinase conformational change (Jmol): http://www.biochem.arizona.edu/classes/bioc462/462a/jmol/hexokinase/newhk.html movie of chemical mechanism of serine proteases (from Voet & Voet, Biochemistry 3rd ed Biochemistry, ed., 2004 2004, Wiley): http://www.biochem.arizona.edu/classes/bioc460/spring/460web/lectures/LEC1314_EnzCatMech/15-3c_SerineProtease-b3/SerineProtease.htm Serine proteases (Jmol) http://www.biochem.arizona.edu/classes/bioc462/462a/jmol/serprot/serprot1.htm Induced Fit: citrate synthase open form closed form Berg et al., Fig. 17-20 Key Concepts • Mechanisms used by enzymes to enhance reaction rates include: (1st 4 mechanisms based on BINDING of substrate and/or transition state) 1. Proximity & orientation 2. Desolvation (one type of electrostatic catalysis) 3. Preferential binding of the transition state 4 Induced fit 4. 5. General acid/base catalysis 6. Covalent (nucleophilic) catalysis 7. Metal ion catalysis 8. (Electrostatic catalysis) • The chemical mechanism of serine proteases like chymotrypsin illustrates: – Proximity and orientation – Transition state stabilization – Covalent catalysis, involving a “catalytic triad” of Asp, His and Ser in the active site – general acid-base catalysis – electrostatic catalysis Enzymes: Catalytic Strategies 1 BIOC 460 Summer 2010 General Catalytic Mechanisms • Different enzymes use different combinations of mechanisms to reduce activation energy (∆G‡) and thus increase rate of reaction. • 7 (or 8) "types" of mechanisms below -- really "overlapping" concepts in many cases. • 1st 4 mechanisms related to BINDING of substrate and/or transition state state, (reaction takes place in active site, not in bulk solution) 1. PROXIMITY AND ORIENTATION (catalysis by “approximation”) – Proximity • Reaction between bound molecules doesn't require an improbable collision of 2 molecules. • They They're re already in "contact" contact (increases local concentration of reactants). – Orientation • Reactants are not only near each other on enzyme, they're oriented in optimal position to react. • The improbability of colliding in correct orientation is taken care of. General Catalytic Mechanisms, continued 2. DESOLVATION • Active site gets reactants out of H2O. • Lower dielectric constant environment than H2O (more nonpolar environment), so stronger electrostatic interactions (strength inversely related to dielectric constant). • Reactive groups of reactants are protected from H2O, so H2O doesn't compete with reactants. – H2O won't react to give unwanted byproducts, e.g., by hydrolysis of some reactive intermediate in the reaction that was supposed to transfer its reactive group to another substrate. 3. TIGHT TRANSITION STATE BINDING • used to be called "strain and distortion" • Enzyme binds transition state very tightly, tighter than substrate (stabilizes T.S.) • Free energy of transition state (peak of free energy barrier on reaction diagram) is lowered because its "distortion" (electrostatic or structural) is "paid for" by tighter binding of transition state than of substrate. Enzymes: Catalytic Strategies 2 BIOC 460 Summer 2010 4. INDUCED FIT • Conformational change resulting from substrate binding • Binding may stabilize different conformation of enzyme or substrate or both. • Conformational change – orients catalytic groups – promotes tighter transition state binding, and/or – excludes H2O • Example: hexokinase, binding of glucose (1st reaction in glycolysis) – closure of enzyme around upon binding of D-glucose (red) – hexokinase conformational change (jmol) “Open” conformation (no glucose) “Closed” conformation (glucose bound) Hexokinase, induced fit Nelson & Cox, Lehninger Principles of Biochemistry, 4th ed. Fig. 8-21 “Open” conformation (no glucose) Nelson & Cox, Lehninger Principles of Biochemistry, 4th ed. Fig. 8-21 Enzymes: Catalytic Strategies 3 BIOC 460 Summer 2010 “Closed” conformation (glucose bound) Nelson & Cox, Lehninger Principles of Biochemistry, 4th ed. Fig. 8-21 1st 4 concepts (above) in catalysis rather general, all related to BINDING of substrate and/or transition state Other catalytic mechanisms (below) involve specific groups and chemical mechanisms that depend on the specific reaction. 5. GENERAL ACID-BASE CATALYSIS • Specific functional groups in enzyme structure positioned to – donate a proton (act as a general acid) acid), or – accept a proton (act as a general base) • helps enzyme avoid unstable charged intermediates in reaction • the general acid (H+ donor) has to then accept a proton (act as a general base) later in catalytic mechanism to regenerate itself. • Likewise, general base that accepts a proton must give it up later. • Amino acid functional groups that can act as general acids/general bases: – thiol of Cys – R group carboxyls of Glu Glu, Asp – ROH of Ser, Thr, and Tyr − α-amino group – His imidazole – guanidino group of Arg − ε-amino group of Lys Enzymes: Catalytic Strategies 4 BIOC 460 Summer 2010 6. COVALENT CATALYSIS • rate acceleration by transient formation of a COVALENT enzymesubstrate bond • Covalent intermediate is more reactive in next step in reaction, so that step has lower activation energy than it would have for a noncovalent catalytic mechanism -- enzyme alters pathway to get to product. • Nucleophilic attack often involved in covalent catalysis • Nucleophile: an electron-rich group that attacks nuclei • unprotonated His imidazole • unprotonated α-amino group • unprotonated ε-amino group of Lys • unprotonated thiol of Cys (thiolate anion, RS–) • Unprotonated alcohol of Ser Ser,Thr, Thr and Tyr (alkoxide (alkoxide, RO-) • unprotonated R group of Glu, Asp (carboxylates, COO- ) – some coenzymes, e.g., thiamine pyrophosphate (TPP) & pyridoxal phosphate (PLP) 7. METAL ION CATALYSIS (several catalytic roles) Metal ions can be – tightly bound (metalloenzymes), i.e., as a prosthetic group (usually transition metal ions, e.g., Fe2+ or Fe3+, Zn2+, Cu2+, Mn2+....) – loosely bound, binding reversibly and dissociating from enzyme (usually Na+, K+, Mg2+, Ca2+, Zn2+...) • Functions of metal ions in catalysis: A Binding and orientation of substrate (ionic interactions with A. negatively charged substrate) B. Redox reactions (e.g., Fe2+ / Fe3+ in some enzymes) C. Shielding or stabilizing negative charges on substrate or on transition state (electrophilic catalysis) – example: Kinases (e.g., hexokinase) bind ATP (adenosine triphosphate) and require Mg2+ to be bound to nucleotide (so ligand is actually Mg2+•ATP) in order to g charges, g and • shield negative • orient the ATP substrate – All kinases require Mg2+ for activity. • Enzymes: Catalytic Strategies 5 BIOC 460 Summer 2010 [8. ELECTROSTATIC EFFECTS] • concept not always "listed" separately because it’s involved in many other aspects of catalytic mechanisms • Some examples: – providing lower dielectric constant of environment in active site (hydrophobic environment) – altering pK values of specific functional groups – stabilizing a particular conformation of critical groups in active site by electrostatic interactions – stabilizing (binding) a charged intermediate or transition state by providing an oppositely charged enzyme group nearby. PROTEASES • Reaction catalyzed = hydrolysis of peptide bonds • Functions in vivo: – digestion of nutrient protein – degradation of unwanted proteins – specific proteolytic cleavage for activation of specific (developmental processes; blood clotting) • Peptide bond hydrolysis (SN2 attack by :O of water on carbonyl C of the peptide bond) + H+ • Equilibrium (in 55.5 M H2O) lies FAR to the right, but in absence of catalyst, reaction is extremely slow; t1/2 = 10 – 1000 yrs. • Peptide bonds "kinetically stable" Enzymes: Catalytic Strategies 6 BIOC 460 Summer 2010 Mammalian Serine Proteases are Homologous. • 3-dimensional folds (tertiary structures) of chymotrypsin (red) and trypsin (blue). • Mammalian serine proteases: chymotrypsin, trypsin & elastase • Vary with respect to Substrate specificity • Homologous (share a common evolutionary ancestral gene) -primary structures about 40% identical and 3-dimensional folds nearly identical • Common evolutionary origin with a single ancestral gene that duplicated a number of times, after which sequences and substrate specificities diverged (example of divergent evolution.) • Also includes many proteolytic enzymes in the blood clotting cascade. Berg et al., Fig. 9-12 Substrate Binding: the hydrophobic “specificity pocket” of chymotrypsin • area of substrate recognition site responsible for specificity • Position of aromatic ring of S bound in pocket is shown. • Note small Gly residues in “lining” of pocket • Also note Ser 189 in bottom of pocket. (This residue is Asp in structure of trypsin) Berg et al., Fig. 9-10 Enzymes: Catalytic Strategies 7 BIOC 460 Summer 2010 Specificity pockets of chymotrypsin, trypsin, and elastase • Substrate binding sites where "R1" group of the substrate binds ("R1" = R group of the amino acid residue contributing the carbonyl group of the peptide bond to be cleaved). • Trypsin cleaves peptide bonds on carbonyl side ("after") long + charged residues (R1 = Lys+ or Arg+) facilitated by Asp– residue in bottom of S1 site. • Pocket of elastase is occluded so only small side chains may enter. Elastase cleaves after small neutral residues (e.g., Gly and Ala). Berg et al., Fig. 9-13 Mechanism of Peptide Bond Hydrolysis • Mechanism of uncatalyzed reaction: + H+ • SN2 Nucleophilic attack by :O of H2O on carbonyl C • Formation of tetrahedral intermediate • Tetrahedral intermediate then breaks down • Partial double bond character of peptide bond makes carbonyl carbon a poor nucleophilic site. • Catalytic task of proteases is to make that normally unreactive carbonyl group more susceptible to nucleophilic attack by H2O. Enzymes: Catalytic Strategies 8 BIOC 460 Summer 2010 4 Classes of Proteases • 4 classes of proteases based on different mechanisms to enhance the susceptibility of the carbonyl group to nucleophilic attack 1. Serine proteases (e.g., chymotrypsin) -- covalent catalysis, with initial nucleophilic attack carried out by enzyme Ser-O(H) group made into a p potent nucleophile p with assistance of nearby y His imidazole that acts as a general base 2. Cys proteases -- again, covalent catalysis, with initial nucleophilic attack carried out by an enzyme Cys-S(H) group using nearby His imidazole as a general base 3. Asp proteases -- nucleophile is HOH itself, assisted by 2 Asp residues, general base catalysis by 1st Asp carboxyl group and orientation/polarization of substrate carbonyl by 2nd Asp residue 4. Metalloproteases -- again, nucleophile is HOH assisted by metal (e.g. Zn2+) and by general base catalysis due to Glu-COO–. The mechanism of protease catalysis is similar to that found in many classes of other enzymes: kinases, phosphatases, transferases. Detailed look at a serine protease, chymotrypsin • Chymotrypsin makes carbonyl C of peptide bond more reactive by changing pathway of reaction. • Covalent catalysis by Ser residue, with assistance of a general base (His) • Overall reaction: 2 separate "half reactions" (2 "phases" of catalysis), with a metastable covalent intermediate (("acyl-enzyme y y intermediate")) between the 2 half reactions. • Overall chemical steps in the 2nd phase are almost an exact repeat of steps in the first phase. What’s an “acyl group”? Enzymes: Catalytic Strategies 9 BIOC 460 Summer 2010 Overview of chymotrypsin mechanism: 2 half reactions • First step/phase ("acylation") – Enzyme provides potent nucleophile, a specific Ser O(H) group. – Ser OH made more nucleophilic than usual with assistance of nearby His residue as general base – Nucleophilic attack --> the acyl enzyme intermediate (covalent) – Amine "half" of original peptide/protein released as product (P1) at end of first phase. phase S1 P1 S2 P2 Berg et al., Fig. 9-5 Overview of chymotrypsin mechanism: 2 half reactions • Second phase ("deacylation") – 2nd substrate, H2O, is nucleophile, attacking carbonyl C of the carboxylate ester of acyl enzyme, again with assistance of active site His residue as general base. – Ester bond of acyl-intermediate acyl intermediate hydrolyzed regenerating alcohol component (the enzyme chymotrypsin, with its Ser-OH free again) and carboxylic acid component, the 2nd product (P2) (carboxyl "half" of original substrate peptide/protein). Berg et al., Fig. 9-5 Enzymes: Catalytic Strategies 10 BIOC 460 Summer 2010 THE CATALYTIC TRIAD • 3 amino acid residues in active site in a hydrogen-bonded network: – Ser 195 – His 57 – Asp 102 • essential ti l for f effective ff ti catalytic t l ti activity ti it in i chymotrypsin h t i • Catalytic triad action converts OH group of Ser 195 into a potent nucleophile. Berg et al., Fig. 9-7 Movie of chemical mechanism of chymotrypsin: http://www.biochem.arizona.edu/classes/bioc460/spring/460web/lectures/LEC1314_EnzCatMech/15-3c_SerineProtease-b3/SerineProtease.htm Whole Chymotrypsin Mechanism (Berg et al., Fig. 9-8) Enzymes: Catalytic Strategies 11 BIOC 460 Summer 2010 FIRST PHASE: ACYLATION of Enzyme • Formation of acyl-enzyme covalent intermediate and generation of the amine product 1. Formation of ES Complex • Enzyme binds substrate with bulky aromatic side chain in "specificity pocket”. • Bound substrate is positioned for the peptide bond on carbonyl side (i.e., "carboxyl" side) of that residue to be cleaved. Berg et al., Fig. 9-8, Step 1 2. Formation of 1st Tetrahedral Intermediate (the chemistry begins) • • • Oxygen atom of active site Ser-OH activated by hydrogen bond to His (imidazole ring N:) in catalytic triad Ser-O(–) carries out nucleophilic attack on carbonyl C of substrate (i.e. covalent catalysis) --> COVALENT bond to carbonyl C (1st tetrahedral intermediate). Asp in catalytic triad: a) helps maintain perfect orientation of His and Ser residues in hydrogen bonded network, and b) facilitates H+ transfer by electrostatic stabilization of HisH+ after it has accepted the proton. Berg et al., Fig. 9-8, Step 2 Enzymes: Catalytic Strategies 12 BIOC 460 Summer 2010 Transition State Stabilization, the “Oxyanion Hole” • FIRST TETRAHEDRAL INTERMEDIATE: similar to that of transition state for its formation and breakdown, with negatively charged OXYANION •The "oxyanion hole", an area in the active site of serine proteases that binds the transition state particularly tightly tightly. •Active site binds oxyanion more tightly than it bound original carbonyl group of the substrate. •C–O- bond longer than C=O •Additional hydrogen bond forms with peptide NH groups •Net result: Stabilization of tetrahedral intermediate Berg et al., Fig. 9.9 3. Formation of Acyl-Enzyme Intermediate • • • First tetrahedral intermediate breaks down -- original amide (peptide) bond cleaves HisH+ donates a proton to the amino "half" of the original substrate (HisH+ now a general acid) to generate R2-NH2. Acyl-enzyme intermediate: Peptide covalently attached to Ser residue of enzyme. Berg et al., Fig. 9-8, Step 3 Enzymes: Catalytic Strategies 13 BIOC 460 Summer 2010 4. Amine Product (R2-NH2) dissociates from active site (1st product leaves). • Original carbonyl group of peptide b d iis now a carbonyl bond b l group again, but it's covalently attached to the Ser-O in the acyl-enzyme product of first half reaction (acylation phase). Berg et al., Fig. 9-8, Step 4 SECOND PHASE OF CATALYSIS: DEACYLATION • Breakdown of acyl-enzyme (covalent intermediate) by reaction with H2O (HYDROLYSIS) and release of the carboxylic acid product • Almost an exact repeat of the first steps in terms of catalytic steps/mechanisms 5. Binding of Second Substrate, H2O, in Active Site • H2O activated by His acting as general base Berg et al., Fig. 9-8, Step 5 Enzymes: Catalytic Strategies 14 BIOC 460 Summer 2010 • 6. Formation of the Second Tetrahedral Intermediate Nucleophilic attack of HO- on carbonyl C of acyl-enzyme intermediate → COVALENT bond between OH and carbonyl C --> 2nd tetrhedral intermediate. Berg et al., Fig. 9-8, Step 6 Transition State Stabilization 2, the “oxyanion hole” again • 2nd TETRAHEDRAL INTERMEDIATE with negatively charged oxyanion. • 2nd TI binding g in oxyanion y hole is similar to binding of first TI. • Oxyanion hole is presumed to be stabilizing transition states for formation and breakdown of 2nd tetrahedral intermediate by binding them tightly. Enzymes: Catalytic Strategies 15 BIOC 460 Summer 2010 7. Breakdown of 2nd Tetrahedral Intermediate: Original ester bond (from acyl-enzyme) CLEAVES. • • HisH+ (general acid) donates proton back to Ser O, generating alcohol product of hydrolysis of acyl-enzyme, Ser-OH Ester bond from acyl-enzyme intermediate breaks --> carboxylic acid product d t (R1-COOH) COOH) ffrom original i i l substrate. b t t Berg et al., Fig. 9-8, Step 7 8. Carboxylic acid product dissociates from active site. • Enzyme molecule now in its original state, with His imidazole in neutral form, catalytic triad appropriately hydrogen-bonded, and active site ready to bind another molecule of substrate and do it all again. Does hydrolysis occur in the acylation or deacylation half reaction of serine proteases? What is the nucleophile in the acylation half reaction? What is the nucleophile in the deacylation half reaction? Berg et al., Fig. 9-8, Step 8 Enzymes: Catalytic Strategies 16 BIOC 460 Summer 2010 Activation strategies for 3 more classes of proteases (besides Ser proteases) • problem faced by proteases: activation of carbonyl C of peptide bond for attack by a nucleophile • All generate a potent nucleophile to attack peptide carbonyl group. – Cys proteases: nucleophile a Cys thiol activated by His (gen. base) – Asp proteases: nucleophile is HOH itself assisted by 2 Asp residues: general base catalysis by 1 Asp carboxyl group and orientation/polarization of substrate carbonyl by 2nd Asp residue – Metalloproteases: nucleophile is HOH assisted by binding to a metal (e.g. Zn2+) and by general base catalysis by some enzyme base group, e.g. Glu-COO–. Berg et al., Fig. 9-18 HIV protease: an Asp protease • Homodimer: 2 identical subunits, each contributing an Asp to active site. • 2 catalytic Asp residues, 1 from each subunit, on opposite sides of 2-fold axis of symmetry (below the bound crixivan in Fig. 9-21). • Structure in Fig. 9.19 has substrate binding pocket indicated, with the 2 catalytic Asp residues in ball-and-stick structures. • "Flaps" (a portion of each polypeptide chain, labeled) close down after ) substrate binds ((induced fit). • Structure shown in Fig. 9.21 is in complex with an inhibitor, crixivan, which has a conformation that approximates the 2-fold symmetry of the enzyme. • Crixivan thus inhibits HIV protease without affecting normal cellular Asp proteases, which don't have the 2-fold symmetry that HIV protease has. • Crixivan designed to mimic tetrahedral intermediate (transition state) -- it's a transition state analog, with groups to bind various sub-pockets in substrate binding site. Berg et al., Fig. 9-19 Enzymes: Catalytic Strategies Berg et al., Fig. 9-21 17 BIOC 460 Summer 2010 Learning Objectives • Discuss (briefly explain): 8 general catalytic mechanisms used by enzymes to increase the rates of chemical reactions. (You won't be asked on an exam to simply LIST them, but you could be expected to explain any one -or 2 or 3 or 4 -- of them.) • Terminology: proteolysis, serine protease, general acid, general base, catalytic triad, acyl group, tetrahedral intermediate, acyl-enzyme i t intermediate, di t acylation, l ti d deacylation, l ti nucleophile, l hil oxyanion i h hole l • Explain why peptide bonds are kinetically stable in the absence of a catalyst, given that equilibrium lies far in the direction of hydrolysis in 55.5 M H2O. (Why is any specific reaction a slow reaction?) • Describe the chemical mechanism of hydrolysis of peptide bonds by chymotrypsin, including the following: – What is the "job" of the catalyst (the protease), i.e., what group needs to be made more susceptible p to nucleophilic p attack? – Describe substrate binding, including the role and chemical nature of the "specificity pocket" in chymotrypsin, and which peptide bond in the substrate (relative to the specificity group) will be cleaved. – Draw the structure of the catalytic triad at the beginning of the reaction, and explain how the states of ionization and hydrogen bonding pattern of those 3 groups change step by step during catalysis. Learning Objectives, continued • (chemical mechanism of chymotrypsin, continued) – Explain the role of each member of the catalytic triad in the reaction. – Identify the nucleophile that attacks the carbonyl carbon in acylation; identify the nucleophile that attacks the carbonyl carbon in deacylation. – Describe the acyl-enzyme intermediate, including identifying the type of bond attaching the acyl group to the enzyme (Is it an amide linkage? anhydride? ester? etc.) and how that acyl group relates to the structure of the original substrate. – Draw the structures of each of the tetrahedral intermediates in the reaction. (If you can do this, you understand the chemistry by which they formed.) – Identify the leaving group coming from each of the tetrahedral intermediates as the intermediate breaks down. – State what is being acylated and deacylated in the chymotrypsin reaction (be specific about the functional group involved). – Explain the role of the "oxyanion hole" in the mechanism. – Describe which type(s) of general catalytic mechanisms (first learning objective above) are used by chymotrypsin, and how. Enzymes: Catalytic Strategies 18 BIOC 460 Summer 2010 Learning Objectives, continued • • • Compare (very briefly, just the “bottom line”) the overall 3-dimensional structures of chymotrypsin, trypsin, and elastase, and compare the substrate binding specificities of those 3 enzymes, explaining the relationship of the “specificity site/pocket” structure to the differences in substrate specificity.. How do 3 other classes of proteases (besides the serine proteases) generate nucleophiles potent enough to attack a peptide carbonyl group? To which protease class does HIV protease belong? Describe the quaternary structure and symmetry of the HIV protease and where in the quaternary structure the active site residues are located. Enzymes: Catalytic Strategies 19