Multicenter Assessment of a Rapid PNA FISH Method for
advertisement

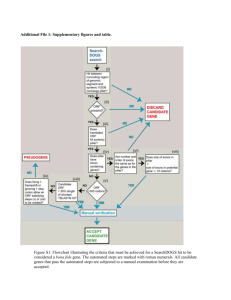
Multicenter Assessment of a Rapid PNA FISH™ Method for the Identification of Candida Species from Blood Culture Bottles C-2073 E. M. Marlowe1, M. Morgan2, S. M. Novak-Weekley1, J. M. Miller1, T. M. Painter3, H. Salimnia3, B. Crystal4 ASM 2010 San Diego, CA 1Southern California Permanente Med. Grp., North Hollywood, CA, 2Cedars Sinai Med. Ctr., Los Angeles, CA, 3Detroit Med. Ctr., Detroit, MI, 4AdvanDx, Woburn, MA. Abstract Candida species are the fourth most common cause of blood stream infections and are associated with significant mortality and health care costs (1,2,3). The C. albicans/C. glabrata PNA FISH™ assay (AdvanDx, Woburn, MA) offers a 2.5 hr result after a blood culture flags positive with the potential of providing important information for species specific treatment. Multicenter studies were undertaken with the objective of evaluating the sensitivity and specificity of a modified, shortened (1.5 hr) procedure for the C. albicans/C. glabrata PNA FISH assay compared to the original (2.5 hr) procedure. This multicenter trial utilized positive blood culture bottles (BACTEC, Becton Dickinson and BacT/ALERT, bioMérieux) which were deidentified clinical discards. A total of 150 samples (125 clinical and 25 seeded) were tested by the C. albicans/C. glabrata PNA FISH assay. There was 100% agreement between the rapid and standard PNA FISH methods as well as with routine identification methods. Specimens included: 61 C. albicans, 31 C. glabrata (including 1 mixed culture positive for C. albicans/C. glabrata) and 59 other yeasts (C. parapsilosis, C. neoformans, C. tropicalis and C. guilliermondii). The new rapid procedure for the C. albicans/C. glabrata PNA FISH provides a faster method for identifying yeast from positive blood culture bottles than the original procedure without effecting the sensitivity or specificity of the assay. The reduced time to results could have an impact on targeted treatment of yeast related blood stream infections. Background Candida species are a common cause of blood stream infections. While C. albicans remains the most commonly isolated, other non-albicans species such as C. glabrata, C. parapsilosis have more recently shown an increased prevalence (2). Appropriate and timely antifungal therapy has been shown to have a significant impact on hospital mortality compared to empiric treatment.(2). Conventional culture schemes often take more than 1 day for species identification. C. albicans/C. glabrata PNA FISH (Peptide Nucleic Acid Fluorescent In-Situ Hybridization) assay (AdvanDx, Woburn, MA) labels specific C. albicans and C. glabrata rRNA identifying these species in approximately 2.5hrs. (Fig.1). The ability to provide even faster results with a Rapid PNA FISH procedure (1.5 hours) may be of even greater utility. Multicenter studies were undertaken with the objective of evaluating the sensitivity and specificity of a modified, rapid procedure for the C.albicans/C. glabrata PNA assay compared to the original procedure and to routine identification methods. Materials and Methods Results Blood Cultures: Blood cultures (clinical discards) submitted to the Southern California Permanente Regional Laboratories, Detroit Medical Center and Cedars Sinai Microbiology Laboratory were used for testing in this study. Blood was inoculated into either; bioMérieux Standard Blood Culture bottles and incubated the BacT/Alert® 3D automated microbial detection system (bioMerieux, Durham, NC), or BACTEC Standard aerobic and anaerobic media and incubated in the BACTEC 9240® automated detection system (BD, Cockeysville, MD). A total of 125 yeast positive blood cultures were included in the study. Standard culture methods as well as the API 20C were used to identify the yeast in all positive blood cultures and compared to the PNA FISH results. Additionally, 25 BACTEC blood culture bottles were seeded with known ATCC strains. PNA FISH: C. albicans/C. glabrata PNA FISH was performed directly from yeast positive blood cultures. On all samples, both the standard FDA cleared procedure and the new rapid method, which reduces the 90 minute hybridization step to 30 minutes and eliminates a 10 ethanol soak, was performed (Fig.2). The operator was blinded to the results of routine identifications and the 2 PNA FISH methods were read independently. Expected are illustrated in figure 3. Figure 1: Illustration of C. albicans/C. glabrata PNA FISH Principle Table 1: Identification of Enrolled Blood Cultures Organism Identified (Routine Methods) Number Table 2: Performance Data for Rapid C. albicans/C. glabrata PNA FISH vs Standard PNA FISH Procedure (results where the same vs. routine identification) C. albicans 46 C. glabrata 27 C. albicans + C. glabrata 1 Study Site C. albicans C. tropicalis 6 A C. neoformans 5 C. parapsilosis 40 C. albicans (ATCC #28815) seeded 14 C. glabrata (ATCC #2001) seeded 3 C. parapsilosis (ATCC #34136) seeded 2 C. guilliermondii (ATCC #6260) seeded 3 C. tropicalis (ATCC # 13803) seeded 1 C. krusei (ATCC # 6258) seeded 2 Total 150 C. glabrata Other Yeast Species Blood Culture System 21/21 9/9 20/20 BACTEC B 21/21 13/13 16/16 BacT/ALERT C 19/191 9/91 23/23 BACTEC Totals Sensitivity 100% (61/61) 95% CI (95.2-100) Sensitivity 100% (31/31) 95% CI (90.8-100) Specificity 100% (59/59) 95% CI (95.1-100) 1One bottle was mixed C. albicans and C. glabrata Summary The Rapid procedure for the C. albicans/C. glabrata PNA FISH provides a faster method for identifying yeast from positive blood culture bottles than the original procedure without effecting the sensitivity or specificity of the assay. Additional studies are needed to determine the impact reduced time to results could have on lab workflow and improved patient outcomes for blood stream infections due to yeast. Figure 3: Fluorescence Microscopy Results Bibliography Left to right: C. albicans, C. glabrata, both C. albicans & C. glabrata, Negative Figure 2: Standard vs. Rapid PNA FISH Protocol X Fixation solution One Drop BC Heat fix 20 min Ethanol 10 min ELIMINATED 1. Hajjeh,R., Sofair, A., Harrison, L., Marshall Lyon, G., Arthington-Skaggs, B., Mirza,S., Phellan, M., Morgan, J., Lee-Y-Yang,W., Ciblak, M., Benjamin, L., Thomson Sanza, L., Huie, S., Fah Yeo, S., Brandt, M., Warnock, D. 2004. Incidence of Bloodstream Infections Due to Candida Species and In Vitro Susceptibilities of Isolates Collected from 1998 to 2000 in a Population-Based active Surveillance Program. Journal of Clinical Microbiology. 42(4):1519-27. 2. Morrell, M., Fraser, V., Kollef, M. 2005. Delaying the Empiric Treatment of Candida Bloodstream Infection until Positive Blood Culture Results Are Obtained: a Potential Risk Factor for Hospital Mortality. Antimicrobial Agents and Chemotherapy. 49(9): 3640-45. Probe/Coverslip Hybridize 90 min REDUCED TO 30 MIN Incubate 30 min Mount coverslip Examine results 3. Zaoutis, T., Argon, J., Chu, J., Berlin, J., Walsh, T., Feudtner, C. The Epidemiology and Attributable Outcomes of Canidemia in Adults and Children Hospitalized in the United States: A Propensity Analysis. Clinical Infectious Disease. 41: 1232-39.