9 Enzyme Inhibition
advertisement

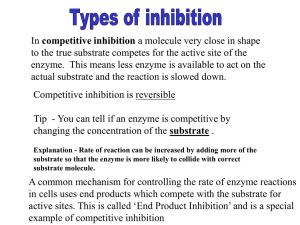
HL IB-Biology 2011/12 #9 ENZYME INHIBITION 7.6 Enzymes Assessment Statements Addressed: 7.6.4 Explain the difference between competitive and non-competitive inhibition, with reference to one example of each. 7.6.5 Explain the control of metabolic pathways by end-product inhibition, including the role of allosteric sites. MECHANISMS OF ENZYME INHIBITION 1. Compare competitive and non-competitive enzyme inhibition. 2. Construct an annotated drawing showing competitive and non-competitive enzyme inhibition. Labudda, CHS, 9 Enzyme Inhibition.pages 3. Sketch how the rate of an enzymatic reaction is changed by the presence of a competitive inhibitor. 4. Sketch how the rate of an enzymatic reaction is changed by the presence of a noncompetitive inhibitor. 5. Explain ‘end-product inhibition’. 6. Show and discuss two examples of end-product inhibition from the control of glycolysis and the isoleucine metabolism in E.coli. 7. State which step is usually inhibited by the end product and provide an explanation. 8. Define allosteric enzyme. 9. State what form of competition is allosteric competition. 10. Outline the role of allosteric enzymes in the control of metabolic pathways. SIMULTING ENZYME INHIBITION WITH TOOTHPICKS DESIGN Research Question What is the effect of an inhibitor on the rate of an enzymatic reaction? Hypothesis In the graph below draw how you predict the rate of the reaction will change in the presence of an enzyme inhibitor. Provide a written explanation for your prediction to the right of the graph. Rat Rate Substrate Variables Independent variable:Substrate concentration (count out needed number of toothpicks), inhibitor concentration (count out needed number of twist ties) Dependent variable: Product concentration (count number of broken toothpicks) Constants: enzyme concentration (2 hands) and time (30 seconds) Control: no inhibitor Groups of two students-> 2 trials per group! Pool your data! ★ ★ ★ ★ Material Two hands (enzyme subunit; thumb and index finger = active site) Toothpicks (substrate) Twist ties (inhibitor) Stopwatch Method 11. RATE OF REACTION DEPENDENT ON SUBSTRATE CONCENTRATION WITHOUT INHIBITOR Time: 30 seconds Substrate concentrations: 10, 20, 35, 50 toothpicks Write out your steps: 12. RATE OF REACTION DEPENDENT ON SUBSTRATE CONCENTRATION WITH INHIBITOR (VARYING CONCENTRATIONS) Time: 30 seconds Substrate concentrations: 10, 20, 35, 50 toothpicks Inhibitor concentrations: 10 and 25 twist ties Write out your steps: DATA COLLECTION AND PROCESSING Raw Data Table(s) Write up a formal DC&P and C&E section Virtual Lab Design First get from me your SL Lab report on enzymes and review the experimental set up. You have available three enzymes, lactase, catecholase and catalase. For each enzyme you know the substrate, products and possible inhibitors. You also have a list of available materials. This list is by no means complete and I expect you to do some research of your own for additional methods and materials needed. Your task is to formulate a research question around enzyme inhibition and to design a series of experiments that will produce data suitable to answer your research question (or questions). An important approach for setting up enzymatic reaction is how to keep the reaction volume constant, while changing other parameters. You also need to consider how to start and stop the reaction, if you want to keep the reaction time constant. We will meet in class to discuss and practice setting up pipetting schemes, so you can communicate your virtual method more effectively. You will also be able to use the Vernier spectrophotometer to see how you can quantify a color change of a solution. System 1: Lactase is an enzyme found in the small intestine of humans. It catalyses the breakdown of the disaccharide lactose to D-Galactose and D-Glucose (figure 1) and it maybe regulated by product inhibition (negative feed-back). ONPG is a chemical analog, which means that it can bind to the active site of lactase. As a result it is converted to Galactose and ONP, which is yellow (absorption 420 nm) (figure 2). Figure 1: The Basic Hydrolysis of the Sugar Lactose by the Enzyme Lactase. http://www.science-projects.com/LacInhib.htm Figure 2: "ONPG" Is a Chemical Analog of the Sugar Lactose and Is Hydrolyzed by the Enzyme Lactase. http://www.science-projects.com/LacInhib.htm System 2: Catecholase is found in some fruits and vegetables. You have seen the effects of this enzyme in the discoloration of apples, peaches, or bananas when they are damaged or peeled (or left halfeaten on a desk or someplace). A related enzyme and substrate produce the dark pigment of hair and skin seen in humans and other animals. Both catecholase and its substrate, catechol, are present in undamaged fruit and potatoes, but the reaction does not begin until the enzyme and its substrate are brought together by cell damage in the presence of oxygen, which promotes the exchange of electrons. In this reaction, oxygen “pulls” electrons from catechol, thus oxidizing catechol and forming a molecule called benzoquinone. The benzoquinone molecules link to form long branched chains that serve as the backbone of a resulting reddish-brown pigment (absorption at 230 nm). The process continues until a complex polymer is formed, similar to the skin pigment melanin. Phenylthiourea (PTU) is an inhibitor of catecholase. Figure 1: The oxidation of catechol. In the presence of catechol oxidase, catechol is converted to benzoquinone. Hydrogens removed from catechol combine with oxygen to form water. System 3: Catalase contains four polypeptide chains, each composed of more than 500 amino acids. This enzyme occurs universally in aerobic organisms. One function of catalase within cells is to prevent the accumulation of toxic levels of hydrogen peroxide (H2O2) formed as a by-product of metabolic processes. The primary reaction catalyzed by catalase is the decomposition of H2O2 to form water and oxygen. 2H2O2 ------> 2H2O + O2 (gas) This reaction occurs spontaneously, but not at a very rapid rate. Catalase speeds up the reaction considerably. Catalase can be inhibited by sodium azide and copper sulphate. Materials: (Not a complete list, do not limit yourself to this, you might need more equipment as you research possible methods.) ❖ test tube racks ❖ small test tubes ❖ calibrated 5-ml pipettes ❖ distilled water ❖ potato extract ❖ catechol ❖ disposable pasteur pipettes with pipette bulb ❖ phenylthiourea (PTU) solution ❖ 3% hydrogen oxide solution ❖ Universal enzyme buffer pH 7.2 ❖ Spectophotometer (range 200 nm to 700 nm) ❖ Cuvettes for spectrophotometer ❖ Vernier gas pressure probe ❖ LabQuest ❖ Sodium azide solution ❖ Copper sulphate solution ❖ Droppers ❖ 0.8% ONPG solution ❖ Lactase solution ❖ Saturated Galactose solution ❖ Saturated Glucose solution ❖ Saturated Sucrose solution ❖ Saturated Lactose solution ❖ Stopwatch PRELIMINARY DESIGN TITLE: DESIGN Research Question -must be focused i.e. relate dependent and independent variable Outline of background and rationale for research question: Hypothesis - state first and then give a logical rationale, i.e. based on what knowledge or observation are you making the prediction Variables - List independent, dependent, and controlled variables. Explain how you will measure the dependent variable and how you will change the independent variable. Explain how you will keep controlled variables constant and/or monitor them. Explain how your control group differ from experimental group(s). Uncertainty - Give values for uncertainty and/or discuss sources for uncertainty and where appropriate how you will address uncertainty in a preliminary experiment!!! Material Diagram - Draw and label a diagram which best shows the technique(s) you will use to measure the dependent variable and to change the independent variable. Show how you will keep controlled variables constant and/or monitor them. Show how control group(s) differ from experimental group(s) - all of the above should also be included in your method. Pipetting scheme: Method - Written description of how you plan to carry out the experiment. Make sure you are specific about, incubation times, set up, buffers, concentrations, etc