Mendelian Genetics page 60
advertisement

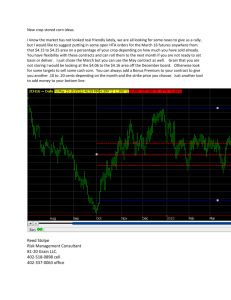
page 60 Mendelian Genetics Objectives 1. Use “genetic corn” to measure and interpret the results of crosses that illustrate Mendel’s laws of segregation and independent assortment. 2. Learn the Chi-square procedure for testing the agreement of observations and expectations, and use this procedure to evaluate the genetic corn results. 3. Demonstrate the principles of ABO and Rho blood typing as an example of codominant inheritance. Procedures 1- Mendelian Genetics of Corn Domestic corn (Zea mays) is ideal for demonstrating the principles of inheritance. An ear of corn is a stem with multiple female flowers arranged along its length. Each flower produces an egg, which will develop into a seed (kernel). The male flowers are on the tassels, which bear anthers and produce pollen. If a young ear of corn on one plant is fertilized with pollen from the tassels of another plant, each kernel of corn that forms on that ear is an individual offspring of the mating. Each ear has hundreds of kernels (offspring), providing the statistical sample that is so important for demonstrating the principles of inheritance. An advantage of domestic corn is that the kernels remain attached to the cob, which makes them easy to observe and handle. Incidentally, the Native Americans that domesticated the corn plant selected an allele that causes the seeds to remain attached to the cob. This is a key mutation that made corn easy to harvest. Respect their accomplishment and DO NOT pick the kernels off the ears! We will examine the phenotypes of the corn kernels and try to explain the observed ratios of phenotypes using Mendel’s principles. You will also perform statistical tests of the results to see if they support the predictions made from Mendel’s principles. First, though, we need to consider just what a corn kernel is and what genes are involved in the tests. Corn kernels consist of several parts (see Figure 1). The outer layer or shell is called the pericarp. The pericarp is not part of the embryo- it is “packaging” from the parent plant. Inside the pericarp is a layer of cells called the aleurone, which can be colored, and the endosperm, which is the bulk of the seed– the starchy part. Also inside is the embryo. We’ll assume that the aleurone, endosperm and the embryo all have the same genotype, which we’ll call this the embryo genotype. We will look at two phenotypic traits- kernel color and kernel shape. In our corn, these characters depend on the embryo genotype. Figure 1– Anatomy of a corn seed (kernel) endosperm Aleurone Pericarp embryo Seed shape. There are two phenotypes in our corn– smooth and wrinkled– and they are controlled by the Su locus. The Su gene has two alleles- these are Su and su. (Try not to be confused by the use of a 2-letter symbol for one allele). Possible diploid genotypes are SuSu, Susu, and susu. Bad enough to write it, but try to say it and make page 61 Mendelian Genetics any sense! Lets call Su starch and su sugar. These alleles control the amount of sugar in the endosperm. The dominant allele Su causes starchy endosperm (little sugar). Homozygous susu produces sugary endosperm (sweet corn) and causes the kernel to be wrinkled and somewhat translucent when it dries. Seed color: The locus that we will consider affects aleurone color and is symbolized Pr. There are two alleles represented in our corn. The allele Pr is dominant and causes the seeds to be dark purple. The allele pr is recessive and causes the kernels to be yellow. Possible genotypes are PrPr, Prpr, and prpr. These two-letter symbols get awkward when you write a genotype for both loci, for example SusuPrPr. Ask your instructor if you don't understand. Table 1. Genetic corn loci and alleles Locus Dominant allele Recessive allele Dominant Effect Su Su su Enzyme converts sugar to starchmakes smooth kernels Pr Pr pr Purple pigment colors kernels (otherwise they are yellow) Count the phenotypes: Work in pairs. The instructor will provide ears resulting from different crosses involving the color and shape genes. Note the code number of each ear and report this number with your results. Examine the ears and be sure that you can distinguish all the phenotypes (smooth vs wrinkled, and purple vs yellow). Use good judgment. For example, a little dimple on the end is not wrinkled- they should be really shriveled. For each ear, count and record the phenotypes of the kernels. Count at least 400 kernels on each ear, or all the kernels, whichever comes first. OK, it is tedious. Doing science is usually 10% inspiration and 90% perspiration. But just think of the glory that was heaped on Mendel (mostly posthumously, of course). Classify each kernel in whole rows, one after the other. When you come to the end of a row, drop down to the next and then work your way back to the other end. As you call out the phenotypes, your partner can keep tally on a piece of scratch paper. Add up the tallies and enter them in Table 2. Note: some ears will have all 4 phenotypes, others only 2. Leave boxes empty as needed. Table 2. Corn phenotype counts ear # Phenotype counts smooth red wrinkled red smooth yellow wrinkled red Total counted page 62 Mendelian Genetics For each ear, calculate the percentages of each phenotype represented (divide the number of kernels X 100 in each category by the total number counted on that ear). Enter the results in Table 3. Table 3. Corn phenotype percentages. ear # Phenotype percentages smooth wrin- smooth red kled red yellow wrinkled yellow 2- Chi-Square Test Remember that inheritance is a matter of probability. How do we decide if our observations fit our predictions? If you were flipping a coin, you would not conclude that the coin was unbalanced just because it came up heads the first two times you flipped it. However, if you flipped heads 9 of 10 times, you would probably be suspicious! Statisticians have quantified these judgments. The Chi-square test is commonly used to compare observed results with predictions according to a specific hypothesis. For example, if you expected 10 of 30 offspring to have the recessive phenotype, and the observed number was 14 of 30, you might want to know how likely it is that the difference between observed and expected result was caused by something other than chance. How different do the observed and expected results have to be before you decide that the prediction is wrong? The formula for calculating the chi-square statistic (X2) is: X 2= Σ[(o-e)2/e] The formula can be stated as follows: chisquare is the sum of the squared differences between observed (o) and expected (e) results in all categories. The symbol Σ means “sum”. What are you summing? The term (o-e)2/e is calculated for each category of the result. For example, the categories in a genetic cross would be each phenotype. For example, let’s say that you expect a 3:1 ratio of phenotypes in a monohybrid test cross. Suppose that the actual results are 638 purple kernels and 240 white, of 878 total. Your hypothesis is the 3:1 ratio, so your prediction is (878*0.75) = 659 purple and (878*0.25) = 219 white. The calculation of X2 for this example is shown in Table 4. Table 4. Calculation of Chi square for the example discussed above. Phenotypes Purple White Observed (o) 638 240 Expected (e) 659 219 (o - e) -21 21 (o-e)2 441 441 (o-e)2/e 0.668 2 X2 = Σ[(o-e)2/e] = 2.668 We get a value of 2.668 for X2. The more the results differ from the predictions, the bigger 2 will be. Statisticians have determined exactly how probable it is to obtain each value of X2 through chance. We can compare our result with a table of X2 values to find its probability. If our results are very unlikely, we can conclude that the prediction was probably wrong. Mendelian Genetics page 63 Of course, we have to pick a criterion for how unlikely a result can be before we reject our hypothesis. The usual standard in natural sciences is 5% (p < 0.05). The criterion is the probability (p) that the result could occur by chance, if the hypothesis is true. If the result would occur less than 5% of the time by chance alone, its fair to guess that a X2 so large is not due to chance*. When we say a result is “statistically significant”, we mean that it is very unlikely to occur by chance alone (< 5% of the time) . Step-by-Step Procedure for Testing Your Hypothesis and Calculating Chi-Square The value of X2 also depends on the number of categories being compared. In this simple example, there are only two phenotypes. Therefore there are only two categories in the calculation of X2. If you think about it, it seems a bit redundant to include both categories in the calculation. After all, if the kernels don’t have the one phenotype, they must have the other. This consideration is called the “degrees of freedom” (df). Degrees of freedom is the number of categories in the data minus 1. In our example, there are two categories (purple and white); therefore, there is 1 degree of freedom. 4. Compare your value of X2 to the chisquare distribution table to determine how likely that value is: Now we are ready to see how unlikely our result really is. Refer to the chi-square distribution table (Table 5 below). Using the appropriate degrees of freedom, locate the value nearest to the calculated value of chisquare. For our example (X2 = 2.67) you should find that the p value is about 0.10. This means that there is a 10% probability that you would see results this far from predictions (or further) due to chance. In terms of your criterion, the results do not differ significantly from expected. *Keep in mind that unlikely results can happen by chance. If you were a real stickler for proof, you might set your criterion at p=0.01 rather than 0.05. 1. State the hypothesis and the predicted results. 2. Based on the hypothesis, predict the expected numbers for each observational class. Remember to use numbers, not percentages. 3. Calculate X2. Round your answer to two decimal places. A. Determine df and locate the appropriate row. B. Locate the value closest to your calculated 2 on that row. C. Read the p value from the head of that column. 5. State your conclusion in terms of your hypothesis. A. If the p value for the calculated X 2 is p > 0.05, accept your hypothesis. 'The deviation is small enough that chance alone accounts for it. A p value of 0.6, for example, means that there is a 60% probability that this X2 or a larger one could occur by chance. The difference between observation and prediction is not significant. B. If the p value for the calculated X2 is p < 0.05, reject your hypothesis, and conclude that something other than chance is operating for the difference to be so large. Chi-square requires that you use numerical values, not percentages or ratios. Also note that Chi-square is not reliable if the expected value in any category is less than 5. page 64 Mendelian Genetics Chi-Square Distribution. Compare values of X2 to this table to determine how likely they are to be due to chance alone. You won’t probably won’t find your exact value of X2. Just check to see if your value is larger or smaller than the X2 for the critical value of p (0.05) and the appropriate degrees of freedom. (df) Probability (p) 0.95 0.90 0.80 0.70 0.50 0.30 0.20 0.10 0.05 0.01 0.001 1 0.004 0.02 0.06 0.15 0.46 1.07 1.64 2.71 3.84 6.64 10.83 2 0.10 0.21 0.45 0.71 1.39 2.41 3.22 4.60 5.99 9.21 13.82 3 0.35 0.58 1.01 1.42 2.37 3.66 4.64 6.25 7.82 11.34 16.27 4 0.71 1.06 1.65 2.20 3.36 4.88 5.99 7.78 9.49 13.28 18.47 5 1.14 1.61 2.34 3.00 4.35 6.06 7.29 9.24 11.07 15.09 20.52 6 1.63 2.20 3.07 3.83 5.35 7.23 8.56 10.64 12.59 16.81 22.46 7 2.17 2.83 3.82 4.67 6.35 8.38 9.80 12.02 14.07 18.48 24.32 8 2.73 3.49 4.59 5.53 7.34 9.52 11.03 13.36 15.51 20.09 26.12 9 3.32 4.17 5.38 6.39 8.34 10.66 12.24 14.68 16.92 21.67 27.88 10 3.94 4.86 6.18 7.27 9.34 11.78 13.44 15.99 18.31 23.21 29.59 “Not significant” “Significant” 3– Blood Typing The ABO and Rh factor blood groups are a good example of codominance– each of two alleles having simultaneous effects on the phenotype regardless of the other’s presence. There are 3 common alleles of the ABO gene locus. They are IA, IB, and i. The codominant IA and IB alleles cause proteins (form A and form B) to be present on the red blood cells. The recessive i allele does not produce a protein. Depending on which combination of these alleles a person inherits, their blood cells may have the following phenotypes: A, B, AB or O. Foreign proteins that enter your body act as antigens, molecules that cause the production of antibodies. Antibodies are proteins made by the immune system that selectively bind to the antigen that induced them. Each antibody molecule bears two binding sites for its antigen, so antibodies can act as a kind of molecular glue that will cause antigen molecules to stick together. All of this becomes an issue if you receive a blood transfusion of cells that bear a protein form that you do not have on your own cells. In that case you produce antibodies that cause the foreign cells to agglutinate, or stick together. A convenient way to determine blood type is to expose a sample of the blood cells to solutions containing the antibodies, and see which antibodies cause agglutination. The page 65 Mendelian Genetics antibody solutions are called antisera because they are the serum portion of the blood of animals that were sensitized by injections of that particular antigen. In the “old days” we used to test our own blood with antisera in the classroom. However, after AIDS became widespread in the 1980’s, we began using synthetic substitutes for blood, out of the prevailing fear of infection. These substitutes work reasonably well. They are particles coated with certain antigens, and they will agglutinate in the presence of antibody like blood cells would. Sure do miss poking fingers to draw blood though... Procedure: You will be provided with several samples of synthetic blood representing different blood types. Obtain a glass slide and lable the ends A and B. Then place a small drop of antiserum A near the A end and a drop of antiserum B at the B end. Next, place small drops of the “blood” that you are testing near to, but not touching, each of the drops of antisera. Use a clean toothpick to mix each pair of drops together. Be sure to use a different toothpick for each pair. Observe the mixed drops for agglutination. The agglutinating “blood” will take on a grainy appearance as the particles clump together. Determine and note the blood type of each of the “persons” whose blood you are testing. Have your instructor check your results and interpretation. For purposes of the lab final, you may be asked to determine the blood type of a sample using this procedure. Assignment: Prepare the following to hand in next time: Genetic Corn & Chi Square statistics 1. For each of the four ears that you examined, hypothesize about the genotypes of the parents. What parental genotypes would you predict would give the ratios of offspring phenotypes that you saw? 2. Test each of your hypotheses using the Chi square test. For each ear, prepare a table in Excel showing the predicted and observed numbers for each phenotype, and the value of X2. Give the approximate p value (from Table 5), and state whether or not the results are consistent with your hypothesis about parental genotypes (i.e., is the p value greater than or less than 0.05?). Blood typing questions 1. Which blood type shows codominance? Explain. 2. Which blood type is the universal donor? 3. Which blood type is the universal acceptor? 4. What is Rh factor? Research this blood type and write a brief essay in your own words explaining its significance. Compare with the ABO blood groups. Why are women who are Rh negative and marry Rh positive men concerned about effects on the health of their second child, but not their first? Genetics jokes Genetics explain why you look like your father and if you don't why you should. Did you know that sterility is hereditary? If your grandfather didn't have children and your father didn't have children, you won't have children either. And by the way– how do you tell the difference between a male chromosome and a female chromosome? You must take down their genes .