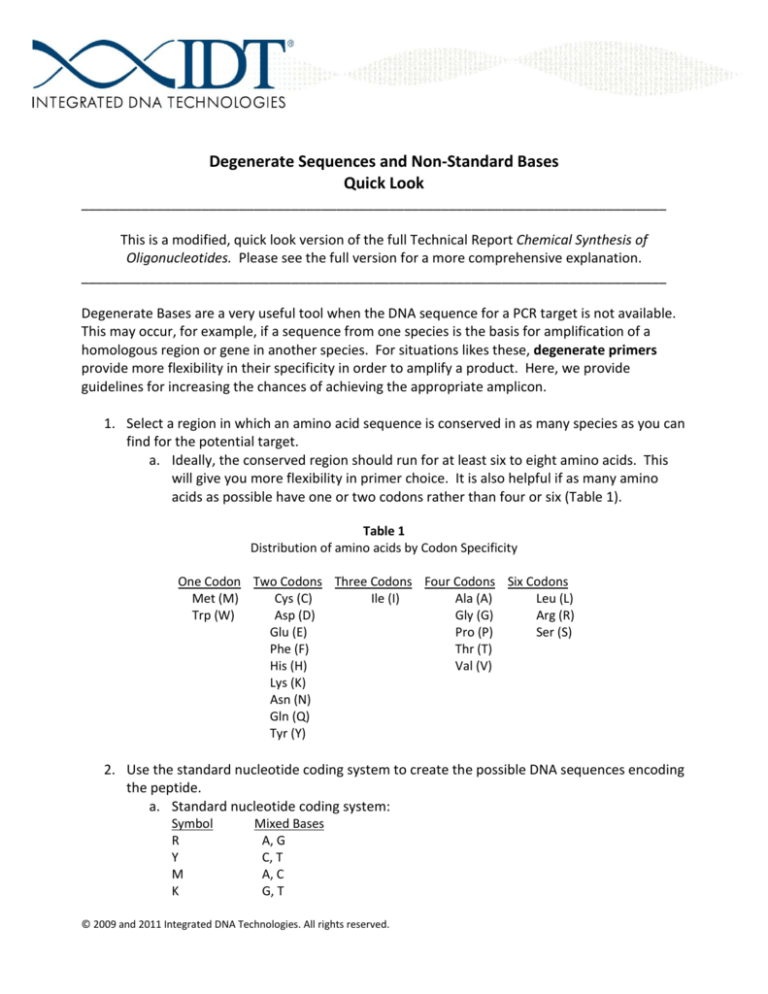
Degenerate Sequences and Non-Standard Bases
Quick Look
______________________________________________________________________________
This is a modified, quick look version of the full Technical Report Chemical Synthesis of
Oligonucleotides. Please see the full version for a more comprehensive explanation.
______________________________________________________________________________
Degenerate Bases are a very useful tool when the DNA sequence for a PCR target is not available.
This may occur, for example, if a sequence from one species is the basis for amplification of a
homologous region or gene in another species. For situations likes these, degenerate primers
provide more flexibility in their specificity in order to amplify a product. Here, we provide
guidelines for increasing the chances of achieving the appropriate amplicon.
1. Select a region in which an amino acid sequence is conserved in as many species as you can
find for the potential target.
a. Ideally, the conserved region should run for at least six to eight amino acids. This
will give you more flexibility in primer choice. It is also helpful if as many amino
acids as possible have one or two codons rather than four or six (Table 1).
Table 1
Distribution of amino acids by Codon Specificity
One Codon Two Codons Three Codons Four Codons Six Codons
Met (M)
Cys (C)
Ile (I)
Ala (A)
Leu (L)
Trp (W)
Asp (D)
Gly (G)
Arg (R)
Glu (E)
Pro (P)
Ser (S)
Phe (F)
Thr (T)
His (H)
Val (V)
Lys (K)
Asn (N)
Gln (Q)
Tyr (Y)
2. Use the standard nucleotide coding system to create the possible DNA sequences encoding
the peptide.
a. Standard nucleotide coding system:
Symbol
R
Y
M
K
Mixed Bases
A, G
C, T
A, C
G, T
© 2009 and 2011 Integrated DNA Technologies. All rights reserved.
S
W
H
B
V
D
N
C, G
A, T
A, C, T
C, G, T
A, C, G
A, G, T
A, C, G, T
b. For example, for the amino acid sequence: GYPVVTCQWD
Gly Tyr Pro Val Val Thr Cys Gln Trp Asp
GGN TAY CCN GTN GTN ACN TGY CAR TGG GAY
3. Make the number of degenerate primers more manageable. This is necessary to reduce
the number of possible degenerate primers. For example, the total number of possible
degenerate primers for the above peptide is 16, 384!
a. Begin by specifying a fixed 3’ end.
i. The last six nucleotides are TGGGAY. Drop off the Y and the last five
nucleotides are fixed as TGGGA.
b. Consult a Codon Usage Table for the species whose DNA is the target. Some species
have a marked preference for certain codons.
i. The best source for this data is the Codon Usage Database
(http://www.kazusa.or.jp/codon/). This site contains thousands of species
listed by their Genus and Species names.
4. An alternative method that can be used along with, or instead of, the mixed base sites
shown above in the universal base approach. Universal bases are analog compounds that
can replace any of the four DNA bases without destabilizing base-pair interactions [1].
a. Commonly used universal bases
Figure 1. Structure of the universal bases 3(Figure 1):
nitropyrrole (1) and 5-nitroindole (2).
i. 3-nitropyrrole
ii. 5-nitroindole
b. The first universal base was 2’deoxyinosine
i. This is still used today but
displays a slight bias in
nucleotide hybridization
with dI:dC being favored
over other pairings [2].
c. For more information on truly
universal base analogs with no pairing bias and no alteration in stability see
reference [3] for a review.
© 2009 and 2011 Integrated DNA Technologies. All rights reserved.
References
1.
2.
3.
Loakes D, Brown DM, et al. (1995) 3-Nitropyrrole and 5-nitroindole as universal bases in
primers for DNA sequencing and PCR. Nucleic Acids Res, 23(13): 2361−2366.
Kawase Y, Iwai S, et al. (1986) Studies on nucleic acid interactions. I. Stabilities of miniduplexes (dG2A4XA4G2-dC2T4YT4C2) and self-complementary d(GGGAAXYTTCCC)
containing deoxyinosine and other mismatched bases. Nucleic Acids Res, 14(19):
7727−7736.
Loakes D. (2001) Survey and summary: The applications of universal DNA base analogues.
Nucleic Acids Res, 29(12): 2437−2447.
© 2009 and 2011 Integrated DNA Technologies. All rights reserved.