CHEM 4590 Practice questions for Test 1 Biological Mass
advertisement
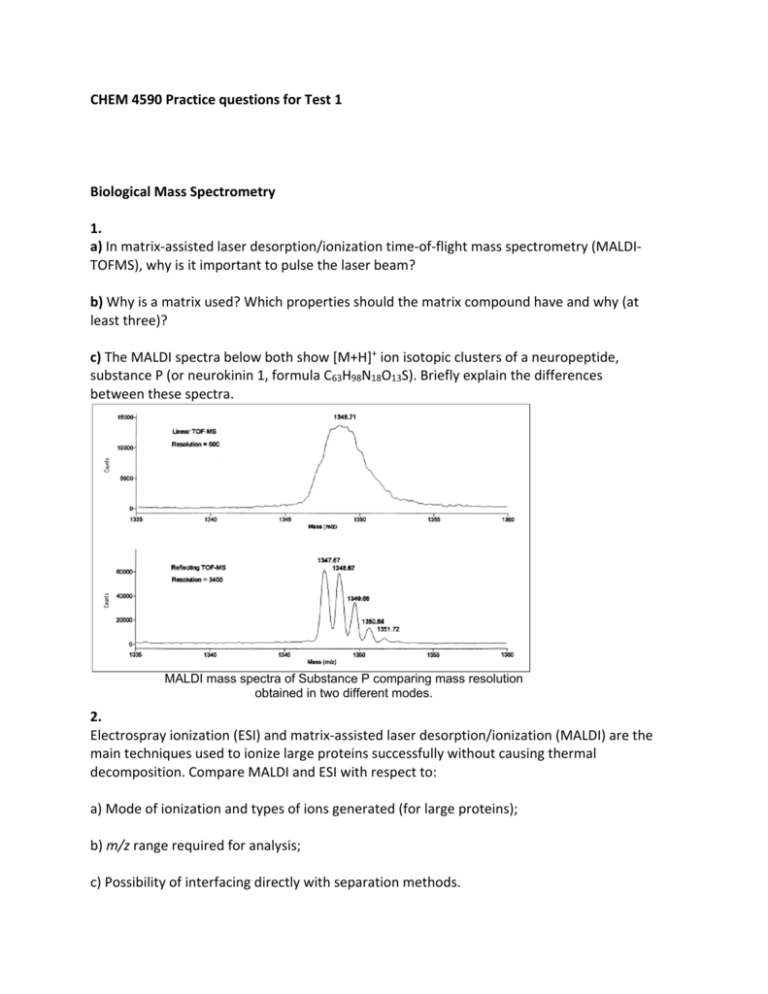
CHEM 4590 Practice questions for Test 1 Biological Mass Spectrometry 1. a) In matrix‐assisted laser desorption/ionization time‐of‐flight mass spectrometry (MALDI‐ TOFMS), why is it important to pulse the laser beam? b) Why is a matrix used? Which properties should the matrix compound have and why (at least three)? c) The MALDI spectra below both show [M+H]+ ion isotopic clusters of a neuropeptide, substance P (or neurokinin 1, formula C63H98N18O13S). Briefly explain the differences between these spectra. MALDI mass spectra of Substance P comparing mass resolution obtained in two different modes. 2. Electrospray ionization (ESI) and matrix‐assisted laser desorption/ionization (MALDI) are the main techniques used to ionize large proteins successfully without causing thermal decomposition. Compare MALDI and ESI with respect to: a) Mode of ionization and types of ions generated (for large proteins); b) m/z range required for analysis; c) Possibility of interfacing directly with separation methods. 3. a) Given a negative mode electrospray ionization (ESI) spectrum where ions are multiply charged by deprotonation (e.g. [M‐nH]n‐), derive a general formula to calculate the charge state (n) sand the molecular mass (M) of an unknown protein, based on measured values of (m/z)1 and (m/z)2. See the spectrum below for reference. Negative mode ESI spectrum (m/z)2 [M-(n+1)H](n+1)- (m/z)1 [M-nH]n- b) Explain why buffers, salts and other impurities should be removed as much as possible from sample solutions prior to electrospray. 4. Calculate the time of flight for an enzyme (protein) of MW 185750 g/mol, if ionized by MALDI, and the ions are accelerated with 15 kV. The TOF tube has a length of 1.5 m, and ions are singly charged. Gel electrophoresis of proteins 1. a) The following standard proteins (shown in table) serve as MW markers during SDS‐PAGE separations. Draw a hypothetical gel showing the migration pattern of these proteins given an optimal gel composition for complete separation. b) What would the separation patterns look like if a lower than optimal % acrylamide was used, or a higher percentage? c) Discuss the effects of post‐translational modifications (phosphorylation, glycosylation) on the mass measurements of proteins by SDS‐PAGE. 2. a) In 2‐D polyacrylamide gel electrophoresis (PAGE), which properties of proteins allow them to be separated in the first and second dimensions? Describe isoelectric focusing. b) Why is sodium dodecyl sulfate (SDS) necessary in one of the dimensions? Justify at least two reasons. c) In SDS PAGE, why is it sometimes necessary to use gels with different percentages of acrylamide? 3. How would you identify and quantify exogenous (porcine) insulin (a high molecular weight protein) in an athlete’s blood? Note: human and porcine insulin molecules have slightly different compositions. 4. What are the roles of the following components in a SDS‐PAGE experiment? DTT; glycerol; EDTA; Tris; a crosslinker. Gel Permeation Chromatography Question 1 a) Describe which types of stationary phases are used in gel permeation chromatography (GPC) of biological molecules, in terms of general composition and properties. b) If you had the choice between gel electrophoresis and gel permeation chromatography to isolate and collect proteins over a high molecular weight range for further processing and analysis, which technique would you choose and why? Question 2 The stationary phases used in GPC (see Question 1) are good supports for affinity chromatography (AC), once chemically modified. What are the main differences between GPC and AC in terms of: a) Nature of interactions between analyte and stationary phase; b) Effect of molecular size on separation; c) Importance of biological function in the separation process. Question 3 a) Which technique among gel permeation chromatography, MALDI‐MS, and SDS‐PAGE is likely to give the most accurate MW information? Which one comes in second order? b) Justify your answer in a) by summarizing the principle behind MW separation for each technique. Question 4 Given the calibration graph below for preparative GPC, which pore size(s) would you select for the separation of a mixture containing proteins of MW 15000, 50000 and 150000 Daltons? Justify. Peptide and protein sequencing Question 1 a) Find the sequence of the peptide from the MS/MS spectrum below, assuming that all ions are either only b or only y. Note: this is not a tryptic peptide. b) This second dimension spectrum was acquired using the monoisotopic (M+H)+ signal as the fragmentation precursor. What would the (M+H)+ isotopic cluster look like in the first dimension? Question 2 Explain how each of mass spectrometry and tandem mass spectrometry (MS/MS) would or would not allow the differentiation of peptides GVFSLK and GLSFVK. In your answer, discuss features observed in the electrospray, MALDI spectra of these peptides and in the MS/MS spectrum of [M+H]+ ions. Use the reference table. Question 3 The peptide PEPTIDE, when measured by mass spectrometry at high resolution (R = 10000), produces an isotope cluster looking like this: a) What would be the m/z value of the monoisotopic (M+H)+ ions? b) These ions, when subjected to collisions with a gas, would produce b and y ions. Determine the m/z values of one series of these fragments (y or b). c) If you turned the resolution down to 500, what types of patterns would you obtain by MS and by MS/MS? General Proteomic approaches 1. Erythropoietin (EPO) is a large protein which helps to enhance the concentration of oxygen in blood. It is present naturally in everyone’s blood, but some athletes choose to use an exogenous form of the protein to supplement their own provision and maintain a higher oxygen level. There are usually minor amino acid sequence differences between the endogenous and exogenous forms. Using some protein analytical tools seen in class, describe how one could: a) purify EPO from blood, b) analyze for detailed structure. 2. If you had to identify and sequence a large protein (e.g. 70 kDa) purified from a cell extract, as quickly as possible, briefly outline a proper strategy (flowchart form) if the tools available are: ‐ Electrospray‐MS and MS/MS mass spectrometer ‐ MALDI‐MS and MS/MS mass spectrometer ‐ GPC column and affinity column ‐ HPLC system with a reversed‐phase (C‐18) column ‐ On‐line protein database search engines ‐ HPLC retention time prediction algorithm ‐ Gel electrophoresis equipment (1‐ and 2D) ‐ Proteolytic enzymes (e.g. trypsin) Note: not every item needs to be used in this procedure. 3. The diagram below shows a possible workflow used in protein analysis and sequencing. In a line or two, explain the purpose of each step, starting with affinity chromatography.