Affinity
advertisement

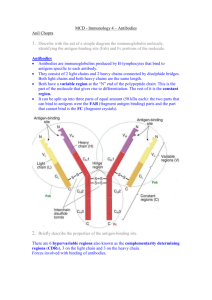
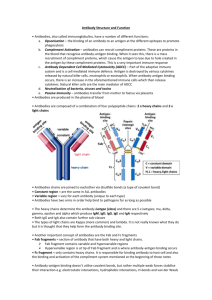
Both antibody and antigen are usually multivalent. Antigens carry multiple epitopes and stimulate production of antibodies which bind to many of them, with different affinities. How well antibody binds antigen is measured by two quantities Affinity: the equilibrium constant of the reaction of Fab fragments of a monoclonal antibody with an antigen containing one copy of epitope to which the antibody binds. Such simple situations are never encountered in the body and almost never in the lab. Avidity: is a “pseudo-equilibrium constant” for a real, complex, antigen carrying many different epitopes, in multiple copies, that binds to real, multivalent,antibody molecules. Affinity • Strength of the reaction between a single antigenic determinant and a single Ab combining site High Affinity Low Affinity Ab Ab Ag Ag Affinity = attractive and repulsive forces Avidity • The overall strength of binding between an Ag with many determinants and multivalent Abs Affinity Avidity Avidity Cross Reactivity • The ability of an individual Ab combining site to react with more than one antigenic determinant. • The ability of a population of Ab molecules to react with more than one Ag Cross reactions Anti-A Ab Anti-A Ab Anti-A Ab Ag A Ag B Ag C Shared epitope Similar epitope Factors Affecting Measurement of Ag/Ab Reactions • Affinity • Avidity Ab excess Ag excess • Ag:Ab ratio Equivalence – Lattice formation A schematic drawing of a typical antibody molecule. Vedi animazione It is composed of four polypeptide chains two identical heavy chains and two identical light chains. The two antigen-binding sites are identical, each formed by the N-terminal region of a light chain and the N-terminal region of a heavy chain. Both the tail (Fc) and hinge region are formed by the two heavy chains. Immunoglobulin domains. The light and heavy chains in an antibody molecule are each folded into repeating domains that are similar to one another. The variable domains (shaded in blue) of the light and heavy chains (VL and VH) make up the antigen-binding sites. The constant domains of the heavy chains (mainly CH2 and CH3) determine the other biological properties of the molecule. Antibody hypervariable regions. Highly schematized drawing of how the three hypervariable regions in each light and heavy chain together form the antigen-binding site of an antibody molecule The folded structure of an IgG antibody molecule, based on x-ray crystallography studies The structure of the whole protein is shown in the middle, while the structure of a constant domain is shown on the left and of a variable domain on the right. Both domains consist of two b sheets, which are joined by a disulfide bond (not shown). Note that all the hypervariable regions (red) form loops at the far end of the variable domain, where they come together to form part of the antigen-binding site Both light and heavy chains are made up of repeating segments each about 110 amino acids long and each containing one intrachain disulfide bond. These repeating segments fold independently to form compact functional units called immunoglobulin (Ig) domains. A light chain consists of one variable (VL) and one constant (CL) domain These domains pair with the variable (VH) and first constant (CH1) domain of the heavy chain to form the antigen-binding region. The remaining constant domains of the heavy chains form the Fc region, which determines the other biological properties of the antibody. The similarity in their domains suggests that antibody chains arose during evolution by a series of gene duplications, beginning with a primordial gene coding for a single 110 amino acid domain of unknown function. A look into the secondary structure Tabular Overview Chains 15C8:H 15C8:L Residues 217 213 Mol. Weight [D] 23064 23224 Chain Type Protein Protein The assignments are: H=helix; B=residue in isolated beta bridge; E=extended beta strand; G=310 helix; I=pi helix; T=hydrogen bonded turn; S=bend. Chain 15C8:H Compound Igg 5C8 Type Protein Molecular Weight 23064 Number of Residues 217 Number of Alpha 0 Content of Alpha 0.00 Number of Beta 20 Content of Beta 47.47 Sequence and secondary structure 1 EVQLQQSGAE LVKPGASVKL SCTASGFNIK DTYMHWVKQK PEQGLEWIAQ EEEE E EE TT EEE EEEEESS GG GSEEEEEEE TTS EEE EE 51 IDPANGNTKY DPKFQGKATI TADTSSNTAY LHLSSLTSED SAVYYCAADP EETTTTEEEE TTTBTTEEE EEETTTTEEE EEE S GGG EEEEEEEE 101 PYYGHGDYWG QGTTLTVSSA KTTPPSVYPL APGSAAQTNS MVTLGCLVKG SSTTS B EEEEE B EEEEE S SSSS EEEEEEEES 151 YFPEPVTVTW NSGSLSSGVH TFPAVLQSDL YTLSSSVTVP SSTWPSETVT EESS EEEE GGGTB SSEE E EESSSS EEEEEEEEE TTTTTTS E 201 CNVAHPASST KVDKKIV EEEEEGGGTE EEEEE The Generation of Antibody Diversity Human antibody preimmune repertoire is made of more than 1012 different antibody molecules • preimmune repertoire low affinity* • repeated stimulation by antigen much higher affinity** How can an animal make more antibodies than there are genes in its genome? (human genome < 50,000 genes) joining separate gene segments together before they are transcribed mammalian immune system almost unlimited number of different light and heavy chains (light and heavy chains of antibodies usually form the antigen-binding site) *the antigen-binding sites of many antibodies can cross-react with a variety of related but different antigenic determinants ** antigen stimulation greatly increases the antibody arsenal The organization of the DNA sequences that encode the constant region of an antibody heavy chain. The coding sequences (exons) for each domain and for the hinge region are separated by noncoding sequences (introns). The intron sequences are removed by splicing the primary RNA transcripts to form mRNA. The presence of introns in the DNA is thought to have facilitated accidental duplications of DNA segments that gave rise to the antibody genes during evolution. Antibodies are produced from three pools of gene segments 1. encodes heavy chains, 2. encodes k light chains, 3. encodes l light chains IgM IgD IgG IgA IgE Catene pesanti m d g a e Catene leggere kol kol kol kol kol % dell’Ig nel sangue 5 <1 80 15 <1 In each pool gene segments that code for different parts of the variable region of the light or heavy chains are brought together by site-specific recombination during B cell development Example: the V-J joining process involved in making a human k light chain During the development of a B cell, the randomly chosen V gene segment (V3 in this case) is moved to lie precisely next to one of the J gene segments (J3 in this Not yet expressed, and therefore not rearranged, antibody genes a cluster of five J gene segments separated from the C-region exon by a short intron and from the 40 V gene segments by thousands of bps. These mRNAs are then translated case). The "extra" J gene segments (J4 and J5) and the intron sequence are transcribed (along with the joined V3 and J3 gene segments and the C-region exon) and then removed by RNA splicing to generate mRNA molecules in which the V3, J3, and C sequences are contiguous. The human heavy-chain gene-segment pool There are 51 V segments, 27 D segments, 6 J segments, and an ordered cluster of C-region exons, each cluster encoding a different class of heavy chain. The D segment (and part of the J segment) encodes amino acids in the most variable part of the V region. The total length of the heavy chain locus is over 2 megabases. Each C region is encoded by multiple exons The genetic mechanisms involved in producing a heavy chain are the same as those shown for light chains except that two DNA rearrangement steps are required instead of one. • First a D segment joins to a J segment • then a V segment joins to the rearranged DJ segment Markers of Non-Self Foreign molecules, too, carry distinctive markers, characteristic shapes called epitopes that protrude from their surfaces. One of the remarkable things about the immune system is its ability to recognize many millions of distinctive non-self molecules, and to respond by producing molecules such as these antibodies—and also cells—that can match and counteract each one of the non-self molecules. Any substance capable of triggering an immune response is known as an antigen. An antigen can be a bacterium or a virus, or even a portion or product of one of these organisms. Tissues or cells from another individual also act as antigens; that's why transplanted tissues are rejected as foreign. Markers of Self At the heart of the immune response is the ability to distinguish between self and nonself. Every body cell carries distinctive molecules that distinguish it as "self." Normally the body's defenses do not attack tissues that carry a self marker; rather, immune cells coexist peaceably with other body cells in a state known as self-tolerance.