Chapter 15 / Lecture Outline 36
advertisement

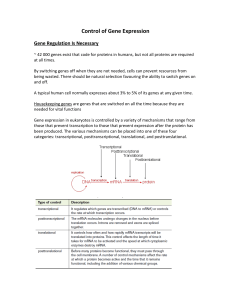
Chapter 15: Gene Regulation in Prokaryotes Outline I. An overview of prokaryotic gene regulation A. Gene expression consists of several steps 1. Details of RNA polymerase form and function in transcription 2. Translation in prokaryotes starts before transcription ends B. The regulation of gene expression can occur at any one of the many steps that transfer information from DNA via RNA to protein II. The regulation of gene transcription A. The utilization of lactose by E. coli: a model system for studying gene regulation 1. The presence of lactose induces expression of the genes required for lactose utilization 2. Analysis of the lactose induction system was a wise choice for the study of gene regulation B. Experiments analyzing the behavior of lactose-utilization mutants reveal the coordinate repression and induction of three genes 1. Complementation analysis of dozens of mutants identifies a set of lactose utilization genes 2. Experimental evidence for a repressor protein 3. How the inducer releases repression to trigger enzyme synthesis 4. The repressor has distinct binding domains – one for the operator and the other for the inducer 5. Changes in the operator DNA to which the repressor binds can affect activity 6. Proteins act in the trans, but DNA sites act only in cis 7. The lac genes are transcribed together C. The operon theory 1. The players 2. The 1961 paper describing the operon theory was a landmark D. A positive control increases transcription of lacZ, lacY, and lacA 1. The CAP protein becomes a positive regulator of the lac and other operons after it forms a complex with cAMP 2. While CAP is a positive regulator for many catabolic operons, some positive regulators increase transcription of the genes in only one pathway E. Summary: how DNA-binding proteins control the initiation of transcription at the lac and other operons F. Molecular studies help fill in the details about control mechanisms 1. Many DNA-binding proteins contain a helix-turn-helix motif 2. Most regulatory proteins are oligomeric and contain more than one binding domain 3. The looping of DNA is a common feature of regulatory systems 4. How regulatory proteins interact with RNA polymerase 5. In studying gene regulation, researchers use the lacZ gene as a reporter of gene expression III. The attenuation of gene expression: fine tuning of the trp operon through the termination of transcription. A. The presence of tryptophan activates a repressor of the trp operon B. The termination of transcription fine-tunes regulation of the trp operon 1. Two alternative transcripts lead to different transcriptional outcomes 2. Simultaneous translation of a small peptide in the leader determines which outcome occurs. IV. Global regulatory mechanisms coordinate the expression of different sets of genes A. An alternate sigma (V) factor mediates E. coli’s global response to heat-shock 1. Studies of mutants unable to generate a heat-shock response helped reveal the mechanisms of global regulation 2. The induction of alternative V factors that recognize different promoter sequences serves as a global regulatory mechanism in many bacteria B. Bacteriophage T7 encodes its own RNA polymerase, which allows the sequential expression of different sets of genes during infection. V. A Comprehensive example: the regulation of virulence genes in V. cholera A. Three regulatory proteins-toxR, toxS, and toxT- turn on the genes for virulence B. A model of virulence regulation leaves some unanswered questions Essential Concepts Most mechanisms of gene regulation in prokaryotes block or enhance the initiation of transcription. In addition, later steps in gene expression are potential targets for fine tuning the amount of gene products that accumulate in cells. In the lac operon model proposed by Jacob and Monod, the binding of the repressor protein (encoded by the lacI gene) to the DNA operator prevents transcription of the structural genes lacZ, lacY and lacA in the absence of the inducer lactose. When lactose is present, its binding to the repressor induces expression of the structural genes by causing the repressor to change its shape and lose its ability to bind to the operator. A critical, general principle emerges from the lac operon studies; Regulatory genes usually encode trans-acting regulatory proteins that interact with cis-acting regulatory DNA elements located near the promoter (such as the operator). Negative regulatory proteins prevent or diminish the rate of transcription, while positive regulatory proteins enhance transcription. Many types of coordinate gene regulation result from the clustering of genes into operons that are transcribed into a single polycistronic mRNA from a single promoter. The binding of repressor proteins to operators can be influenced by either inducers (as for the lac repressor) or co-repressors (as for the trp operon). Catabolite repression regulates certain catabolite operons by preventing the CAP protein, a positive regulator, form binding to the operon’s promoter region, in the presence of high concentrations of glucose. Many regulatory proteins, both positive and negative, contain a helix-turn-helix- motif, function as oliogomers that bind to more than one DNA site, and interact with RNA polymerase to prevent or assist its function. Attenuation, a form of fine tuning for operons involved in the biosynthesis of amino acids, is based on the amount of premature termination of mRNA transcription. The termination, in turn, is determined by the intracellular concentration of tRNAs charged with the amino acid produced by the enzyme products of the structural genes in the operon. Cells can express different sets of genes at different times or under different conditions by using alternate sigma factors or by producing novel RNA polymerases that recognize different classes of promoters.