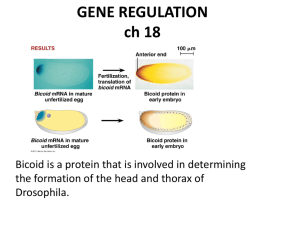
DNA/Gene Regulation
advertisement

Regulation of Gene Expression--AP Biology Prokaryotes A bacterium can adjust its metabolism to the changing environment and available food sources This metabolic control occurs on two levels: Increasing or decreasing the activity of metabolic enzymes Regulating genes that code for metabolic enzymes Operons: The Basic Concept In bacteria, genes are often clustered into operons, composed of An operator--the “on-off” switch A promoter Genes for metabolic enzymes An operon can be switched off by a protein called a repressor A co-repressor is a small molecule that works with a repressor to switch an operon off Repressible and Inducible Operons A repressible operon is one that is usually on; binding of a repressor to the operator shuts off transcription (ex: trp operon) Mutations in repressible operons cause the genes to be continuously transcribed and expressed An inducible operon is one that is usually off (ex: lac operon); a molecule called an inducer inactivates the repressor and turns on transcription Inducible operon lac operon--contains genes coding for enzymes in hydrolysis and metabolism of lactose If lactose is present in the bacterial cells the gene is turned on to make the enzymes that metabolize lactose; the pathway is switched on by a form of lactose Lactose absent, repressor active, operon off Lactose present, repressor inactive, operon on Inducible enzymes usually function in catabolic pathways Repressible enzymes usually function in anabolic pathways Regulation of the trp and lac operons involves negative control of genes because operons are switched off by the active form of the repressor Control of eukaryotic gene expression Typical eukaryotic genome is much larger than that of a prokaryotic cell Gene expression— genes that are available for transcription and translation in a cell Leads to cell differentiation —cells becoming physically different and able to do different functions even though they all have the same DNA DNA organization Proteins called histones are responsible for the first level of DNA packing in chromatin Each “bead” is a nucleosome, the basic unit of DNA packing Nucleosomes supercoil then loop and condense to form chromosomes during cell division Interphase chromatin is usually much less condensed than that of mitotic chromosomes and have: highly condensed areas called heterochromatin, which doesn’t get transcribed and less compacted areas, called euchromatin which do get transcribed Gene expression in eukaryotes All organisms must regulate which genes are expressed at any given time Many key stages of gene expression can be regulated in eukaryotic cells including transcription, translation, and post-translation Is a complex processes that involves Regulatory genes and elements Transcription factors Chromatin modifications Histone acetylation —promotes the initiation of transcription DNA methylation --reduces transcription or causes long-term inactivation of genes in cellular differentiation Regulatory genes and sequences Regulatory gene: sequence of DNA that codes for a regulatory protein or RNA molecule Regulatory sequence: stretches of DNA that interact with regulatory proteins to control transcription Small regulatory RNA’s —small pieces of RNA that can bind to mRNA to block it’s translation Transcription factors Segments of noncoding DNA that help regulate transcription by binding certain proteins called transcription factors Promoters—site where RNA polymerase binds to start transcription Proximal control elements are located close to the promoter Distal control elements, groups of which are called enhancers, may be far away from a gene or even in an intron An activator is a protein that binds to an enhancer and stimulates transcription of a gene Repressors inhibit expression of a particular gene Interactions of all these factors determine how much of the gene product will be produced Gene regulation accounts for some of the phenotypic difference in organisms with similar genomes Mechanisms of Post-Transcriptional Regulation Many types of regulatory mechanisms operate at various stages after transcription Such mechanisms allow a cell to fine-tune gene expression rapidly in response to environmental changes Ex: alternative RNA splicing, different mRNA molecules are produced from the same primary transcript, depending on which RNA segments become exons and which become introns