2 - Springer Static Content Server
advertisement

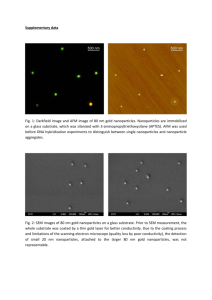
Electrosprayed microparticles with loaded pDNA-calcium phosphate nanoparticles to promote the regeneration of mature blood vessels Xueqin Guo,a Tian Xia,b Huan Wang,a Fang Chen,a Rong Cheng,b Xiaoming Luo,a Xiaohong Lia,* a Key Laboratory of Advanced Technologies of Materials, Ministry of Education of China, School of Materials Science and Engineering, Southwest Jiaotong University, Chengdu 610031, P.R. China b Department of Pathology, The 452nd Hospital of People’s Liberation Army, Chengdu 610021, P.R. China * Corresponding author. School of Materials Science and Engineering, Southwest Jiaotong University, Chengdu 610031, P.R. China. Tel.: +86 28 87634068; fax: +86 28 87634649. E-mail address: xhli@swjtu.edu.cn. S1 Supplementary Materials S1. Preparation of pDNA-loaded nanoparticles with different Ca/P ratios The pDNA-loaded calcium phosphate (CP-pDNA) nanoparticles with Ca/P mole ratios of 1/1, 4/3, and 5/3 were prepared as described previously with some modifications (1). Briefly, 0.5 ml Ca(NO3)2 aqueous solution containing 250 μg pDNA was emulsified into 15 ml cyclohexane containing 10% (v/v) OP-10 by continuous stirring for 1 h to form microemulsion A. Similarly, microemulsion B was formed by emulsification of 0.5 ml (NH4)2HP04 aqueous solution containing 250 μg pDNA into 15 ml cyclohexane containing 10% (v/v) OP-10. Ca(NO3)2 solutions of 0.1, 0.18, and 0.25 M, and (NH4)2HP04 solutions of 0.1, 0.12, and 0.15 M were used to achieve Ca/P ratios of 1/1, 4/3, and 5/3, respectively. Microemulsion B was slowly added to microemulsion A at the rate of 5 ml/h with continuous stirring. The mixture was adjusted to pH 6.0, 6.0, and 9.0 to obtain CP-pDNA nanoparticles with Ca/P mole ratios of 1/1, 4/3, and 5/3, respectively. S2. Characterization of CP-pDNA nanoparticles The particle size and Zeta potential of CP-pDNA nanoparticles were measured by a Nano-ZS laser particle analyzer (Zetasizer Nano ZS, Malvern Co. England). To determine the pDNA encapsulation efficiency, CP-pDNA nanoparticles were extracted three times with phosphate buffer solution (PBS) containing 10 mg/ml heparin. The extract was mixed with Hoechst 33258 and the fluorescence intensity was measured by a fluorospectrophotometer (Hitachi F-7000, Japan) at an excitation wavelength of 350 nm and an emission wavelength of 450 nm. The pDNA concentration was obtained using a standard curve from known concentrations of pDNA solutions. The extraction efficiency was calibrated by adding a certain amount of pDNA solution into blank CP nanoparticles solution along with the same concentration as above and extracted using the above-mentioned process. The encapsulation efficiency indicated the percentage of pDNA encapsulated with respect to the total amount used for the nanoparticle preparation. Table S1 summarizes the size, Zeta potential, pDNA loading efficiency of CP-pDNA nanoparticles prepared under different Ca/P ratios. S2 Table S1. Characterization of CP-pDNA nanoparticles prepared under different Ca/P ratios No. Ca/P ratio pH Diameter (nm) Zeta potential (mV) Encapsulation efficiency (%) 1 5/3 9.0 111 14 -10.9 0.7 89.9 2.4 2 4/3 6.0 115 16 -15.9 0.4 84.3 3.9 3 1/1 6.0 102 15 -12.8 0.3 82.3 3.1 To investigate the crystalline phase of calcium phosphate formed under different conditions, CP-pDNA nanoparticles were analyzed using X-ray powder diffraction (XRD, Philips X’Pert PRO, Philips, The Netherlands) employing Cu Ka radiation (30 kV, 10 mA). The diffraction patterns were collected with 2θ ranging from 20° to 60° with a scanning speed of 0.35°/min. As shown in Fig. S1, hydroxyapatite (HA), octacalcium phosphate (OCP) and dicalcium phosphate dihydrate (DCPD) were formed in CP-pDNA nanoparticles through adjusting the Ca/P feeding ratios and pH values of the precipitation reaction. Fig. S1. XRD profiles CP-pDNA nanoparticles under Ca/P feeding ratios 5/3 (a), 4/3 (b) and 1/1 (c) and reaction media of pH 9.0 (a) and 6.0 (b, c). S3. In vitro cell viability and transfection efficiency of CP-pDNA nanoparticles Human umbilical vein endothelial cells (ECs) and human aortic smooth muscle cells (SMCs) were S3 used to examine the cell viability and gene transfection efficiency. These cells were from American Type Culture Collection (Rockville, MD) and maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco BRL, Rockville, MD) supplemented with 10% heat inactivated fetal bovine serum (FBS, Gibco BRL, Grand Island, NY). CP-pDNA nanoparticles with Ca/P mole ratios of 1/1, 4/3, and 5/3 were tested, using pDNA polyplexes with poly(ethylene imine) (PEI) as control. The pDNA-PEI polyplexes was formed by mixing 500 l of PEI solution (22.3 mM in pH 7.0 PBS) and 500 l of pDNA solution (1 mg/ml in pH 7.0 PBS) (2). The mean diameter of pDNA polyplexes was 278 ± 42 nm, and the zeta potential was 4.1 ± 0.4 mV. EC or SMC suspensions with a cell density of 5 × 104 cells/ml was seeded in 24-well tissue culture plates (TCP), and incubated for 24 h to allow cell attachment. Cells were treated with CP-pDNA nanoparticles and PEI-pDNA polyplexes containing 1.0 μg pDNA. After incubation for 48 h, cell viability was assayed with cell counting kit (CCK8, Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Briefly, after removal of microparticles-containing media, 400 l of fresh culture medium was added in each well. Then 40 l of CCK-8 was added to each well and incubated for 2 h according to the reagent instruction. An aliquot (150 l) of incubation medium was pipetted into a 96-well TCP, and the absorbance at 450 nm was measured for each well using a Quant microplate spectrophotometer (Elx-800, Bio-Tek Instrument Inc., Winooski, VT). All experiments were performed with n = 6. To measure the transfection efficiency, the culture media were removed after 48 h treatment, and cells were homogenized in RIPA lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China). The lysate was centrifuged at 12,000 rpm for 5 min at 4 °C to collect the supernatant. The relative light unit (RLU) was quantified using a fluorospectrophotometer at an excitation wavelength of 493 nm and an emission wavelength of 510 nm. The total protein of the cell lysate was determined by BCA protein assay kit (Pierce, Rockford, IL). Results of pDNA expression were normalized as relative light units per mg of soluble protein (RLU/mg protein). All experiments were performed with n = 6. Table S2 summaries the cell viability and transfection levels of HUVECs and HUVSMCs after incubation with CP-pDNA nanoparticles. S4 Table S2. The cell viability and transfection levels of ECs and SMCs. Sample Cell viability (%) Transfection efficiency (RLU/mg protein×105) EC SMC EC SMC HA-pDNA 93.5 ± 3.1 95.6 ± 3.0 1.27 ± 0.18 0.88 ± 0.07 OCP-pDNA 92.6 ± 3.6 91.9 ± 3.5 0.63 ± 0.09 0.75 ± 0.05 DPCD-pDNA 90.5 ± 3.6 90.0 ± 3.0 0.63 ± 0.13 0.75 ± 0.07 PEI-pDNA 55.0 ± 2.7 55.7 ± 2.9 2.29 ± 0.41 1.88 ± 0.19 References: 1. Morgan TT, Muddana HS, Altinoglu EI, Rouse SM, Tabakovic A, Tabouillot T, et al. Encapsulation of organic molecules in calcium phosphate nanocomposite particles for intracellular imaging and drug delivery. Nano Lett. 2008;8:4108–4115. 2. He SH, Xia T, Wang H, Wei L, Luo XM, Li XH. Multiple releases of polyplexes of plasmids VEGF and bFGF from electrospun fibrous scaffolds towards regeneration of mature blood vessels. Acta Biomater. 2012;8:2659–2669. S5