Validation of a real time-PCR for the detection of Hepatitis B
advertisement

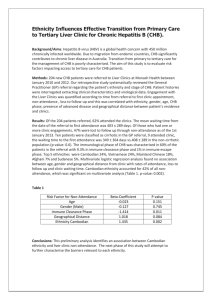
Detection of Hepatitis B covalently closed circular DNA in paraffin-embedded liver biopsy specimens of chronic hepatitis B patients. R.B. Takkenberg1, H.L. Zaaijer2, S. Menting2, C.J. Weegink1, V. Terpstra3, M.G.W. Dijkgraaf4 , M. Cornelissen5, P.L.M. Jansen1, H.W. Reesink1 and M.G.H.M.Beld2 1 AMC Liver Center, Department of Gastroenterology and Hepatology, Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands. 2 Section of Clinical Virology, Department of medical Microbiology, Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands. 3 Department of Pathology, Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands. 4 Department of Clinical Epidemiology and Biostatistics, Academic Medical Center, University of Amsterdam, The Netherlands. 5 Laboratory of Experimental Virology, Department of Medical Microbiology, Academic Medical Center, University of Amsterdam, The Netherlands. Corresponce to: R.B. Takkenberg Meibergdreef 9 Room C2-331 1105 AZ Amsterdam The Netherlands Tel: +3120-5668278 e-mail: r.b.takkenberg@amc.nl 1 Abstract: Introduction: In chronic hepatitis B (CHB) infection, covalently closed circular DNA (cccDNA) is responsible for persistence of infection. No studies, in search of the role of cccDNA, have been performed on formalin-fixed, paraffin embedded (FFPE) liver biopsies. The aim of this study was to detect and analyze cccDNA in FFPE liver tissue and compare the results with individual plasma ALT levels and the Histology Activity Index (HAI). Material and Methods: As we previously described, cccDNA was quantified by using selective primers that target the gap region between the two Direct Repeat Regions (DR1 and DR2). To quantify cells, we used primers that target the GAPDH gene. Real-time PCR (rt PCR) was performed in the Roche Lightcycler® 480. The number of hepatocytes was calculated as 70% of the total number of cells. From 56 FFPE biopsies of HBeAg positive and HBeAg negative CHB patients (29 HBeAg positive and 31 HBeAg negative CHB patients), 60m was cut in coupes of 10m each. Paraffin was extracted with 1 mL xyleen, followed by 2 washes with 100% alcohol and one wash with acetone. DNA extraction was performed according the Boom procedure. Results: The median number of hepatocytes was 9.8x10e6 cells per biopsy (range 2.2x10e64.0x10e7 cells) and cccDNA was detectable in 40 FFPE liver biopsies. The median level of cccDNA/10e6 hepatocytes was significantly higher in biopsies from HBeAg positive patients than HBeAg negative patients (183 copies (range 74-755 copies) vs 23 copies (range 8.0-101 copies) (p=0.0013)). The median level of cccDNA was significantly higher in HBeAg positive CHB patients than in HBeAg negative CHB patients (p=0.0036) and significantly higher in CHB patients with active hepatitis (ALT > 45 U/L) than in CHB patients with inactive hepatitis (ALT ≤ 45 U/L) (p=0.0038). Overall there is a significant correlation between the level of cccDNA/10e6 hepatocytes and viral load (p<0.0001) (correlation 2 coefficient (R) = 0.6). There was no correlation between the level of cccDNA/10e6 hepatocytes and HAI-score (p=0.10). Conclusions: We developed a new rt-PCR for detection of cccDNA in FFPE liver biopsies of CHB patients and showed a strong correlation between the number of cccDNA and viral load. Our findings suggest that FFPE liver biopsies can be used for detection of cccDNA, and that the level of cccDNA in FFPE liver biopsies of CHB patients is indicative of viral replication activity. 3