Wolbachia transmission patterns in different species of Nasonia
advertisement

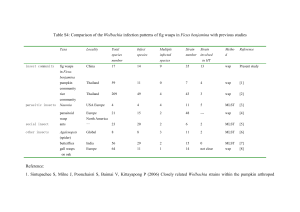
Rachel Martin Marine Biological Laboratory Summer Envisionship 2008 July 30, 2008 Wolbachia transmission patterns in different species of Nasonia Abstract The bacterium, Wolbachia, infects 60-85% of the world’s insect populations by harboring itself in insect gonads. It traditionally has a maternal inheritance pattern. The purpose of this project is to determine if Wolbachia from an infected male was being horizontally transferred to an uninfected female during mating. A cross between an infected male and uninfected female usually results in embryonic death and remains incompatible. This theory was tested in three different species of Nasonia: Nasonia vitripennis, Nasonia longicornis, and Nasonia giraulti. Each species harbors a different type of Wolbachia ranging from high density to low density. Also, offspring from int13.2 (uninfected) females and int12.1 (infected) males, a cross that has previously displayed a horizontal transmission of Wolbachia, were screened to determine if the transmission of Wolbachia was being inherited by the next generation. Uninfected and infected Nasonia were collected before they emerge and separated into tubes to ensure each Nasonia would only mate once. The uninfected females were mated to infected males and then stored in an -80°C freezer until DNA extraction and amplification could proceed. Wsp primers were used in all PCR reactions. The results from these experiments suggest that transmission of Wolbachia does not occur with Nasonia giraulti or Nasonia longicornis. A small rate of transmission was seen in Nasonia vitripennis. The transmission of Wolbachia seen in the cross between int13.2 females and int12.1 males is not inherited in the next generation, as examined in the embryos and diapause. More experimentation will be conducted to expand on these results. Similar methods will be used with Drosophila melanogaster to determine if these results are seen in a different species. Introduction Wolbachia are bacteria that infect a wide range of mainly arthropod hosts. The bacteria are maternally inherited and manipulate host reproduction in order to increase transmission rates in the population. The bacteria cause feminization, male killing, parthenogenesis and cytoplasmic incompatibility (CI) as means of manipulation. More specifically, CI is a form of postfertilization reproductive failure and results in an incompatible cross (Clark et al. 2002). Not all strains of Wolbachia can induce CI. But, the mechanism by which Wolbachia manipulates their host’s reproductive potential, successfully, is unknown. It has been proposed that the bacteria could modify infected sperm during the maturation process. This modification renders them unable to successfully fertilize uninfected eggs (Riparbelli et al. 2007). Three strains of Nasonia have been used to study Wolbachia-induced CI: Nasonia vitripennis, Nasonia longicornis, and Nasonia giraulti. N. giraulti and N. longicornis mostly show embryonic death. In the CI embryos of these two species, the paternal genome does not properly divide to the two daughter nuclei. The severe abnormalities in this segregation result in defects in other mitotic divisions (Tram et al. 2006). In contrast, N. vitripennis show a conversion of the CI embryos to developing males. This is due to the fact that unfertilized eggs normally develop into males. The exclusion of the paternal genome leads to the development of all male offspring (Tram et al. 2006). According to research conducted by Lassy and Karr in 1996, the sperm successfully enters the egg in an incompatible cross. The paternal chromosomes do not properly condense and fuse with the chromosomes of the mother before the first division during mitosis (Clark et al. 2002). In this experiment to determine whether Wolbachia is horizontally transmitted from infected males to uninfected females, I think that the three species of Nasonia will show a Wolbachia infection when mated. The research is beneficial in order to contribute to the knowledge of the biology of Wolbachia. It is helpful to realize how Wolbachia manipulates its hosts. There is not much known about the mechanisms of how Wolbachia cause CI or does not cause CI. Various research can be done to understand more about Wolbachia, which harbors itself in most of the arthropods that inhabit our planet. Methods Mating female Nasonia for DNA Extractions: 1. Crack open hosts a day or two before the Nasonia are due to emerge. 2. Collect black pupae from inside the host. 3. Examine pupae under a microscope to determine the gender of the Nasonia. 4. Separate males and females into different, labeled, glass tubes. 5. Wait for Nasonia to emerge. 6. Put one male and one female into an upside-down glass tube. 7. Watch to guarantee that the Nasonia courted and mated. 8. Remove the female and place into a 1.5ml microcentrifuge tube. 9. Repeat steps 6-8 until all crosses are completed. 10. Place all tubes into a cardboard freezer box and put into -80°C freezer. DNA Extraction of Nasonia Tissue using the Gentra Puregene Tissue Kit: 1. Preheat water baths to 55°C and 65°C. 2. Remove Nasonia from -80°C freezer and put in bench top cooler. 3. Freeze Nasonia in liquid nitrogen. 4. Grind frozen tissue with a pestle. Work quickly and keep tissue on ice at all times. 5. Dispense 300l of Cell Lysis Solution into the 1.5ml grinder tube with the ground tissue. 6. Heat at 65°C for 1 hour. 7. Incubate for 1 minute on ice to quickly cool the sample. 8. Add 100l Protein Precipitation Solution. 9. Vortex vigorously for 20s at high speed. 10. Centrifuge for 10 minutes at 16,000 g. The precipitated proteins should form a tight pellet. If the protein pellet is not tight, incubate on ice for 5 minutes and repeat the centrifugation. 11. Pipet 300l 100% isopropanol into a clean 1.5ml microcentrifuge tube. 12. Add the supernatant from the previous step by pouring carefully. Be careful the protein pellet is not dislodged during pouring. 13. Mix by inverting gently 50 times. 14. Centrifuge 1 min at 16,000g. 15. Carefully discard the supernatant, and drain the tube by inverting on a clean paper towel, taking care that the pellet remains in the tube. 16. Add 300l 70% ethanol and invert several times to wash the DNA pellet. 17. Centrifuge for 1 minute at 16,000g. 18. Carefully discard the supernatant. Drain the tube on a clean paper towel, taking care that the pellet remains in the tube. Allow to air dry for up to 15 minutes. The pellet might be soft and easily dislodged. 19. Add 50l DNA Hydration Solution and vortex for 5 seconds at medium speed to mix. 20. Incubate at 65°C for 1 hour to dissolve the DNA. Samples can be incubated at room temperature overnight with gentle shaking, instead. Ensure tube cap is tightly closed to avoid leakage. DNA Extraction of Nasonia Embryo Tissue using the Gentra Puregene Tissue Kit: 1. Preheat water bath to 65°C. 2. Remove ethanol from embryos using a P20. Be careful not to dislodge embryos from the sides of the tube. They will fit up the P20 tip. 3. Rinse with distilled water. 4. Remove the water. 5. Crush embryos with a pestle. 6. Add 100l Cell Lysis Solution to the embryos and grind gently with a pestle. 7. Incubate at 65°C for 30 minutes. 8. Cool samples to room temperature by placing on ice or putting in fridge for 1 minute. 9. Add 33l Protein Precipitation Solution to cell lysate. 10. Vortex vigorously at high speed for 20 seconds to mix the Protein Precipitation Solution uniformly with the cell lysate. 11. Centrifuge at 16000g for 3 minutes. The precipitated proteins should form a tight pellet. If a tight pellet does not form, repeat sep 3 followed by incubation on ice for 5 minutes and repeat step 4. 12. Pour the supernatant containing the DNA into a clean 1.5ml tube containing 100l 100% Isopropanol. 13. Mix sample gently by inverting several times. 14. Centrifuge at 16000g for 1 minute. The DNA should be visible as a small white pellet. 15. Pour off supernatant and drain the tube on a clean paper towel. Add 100l of 70% ethanol. Invert tube 10 times to wash the DNA. 16. Centrifuge at 16000g for 1 minute. Carefully pour off the supernatant. Pellet may be loose so watch the pellet carefully and pour slowly. 17. Invert and drain on clean paper towel for 10 minutes. 18. Add 20l DNA Hydration Solution to all samples. 19. Rehydrate DNA by incubating sample at 65°C for 1 hour or overnight at room temperature. 20. Store at 4°C. Polymerase Chain Reaction (PCR): 1. Pick a primer to use. This primer should amplify a region specific to what you are looking for. In this case, a Wolbachia surface protein (wsp) primer was used to amplify a region that is unique to Wolbachia. 2. Remove reagents from freezer and place on ice to thaw. 3. Place PCR (.2ml) tubes in a PCR tube tray holder. Label PCR tubes. 4. Create PCR template on Microsoft Excel. 5. Vortex reagents to mix thoroughly. Aliquot reagents into a 1.5l microcentrifuge tube. This mixture is called the “Master Mix.” 6. Place PCR tray with tubes on ice and pipet 9l “ Master Mix” into each PCR tube. When setting up PCR, use specified PCR pipets and filtered pipet tips. 8. Pipet 1l template DNA into the specified PCR tube. 9. Centrifuge PCR tubes and place PCR tubes into a thermocycler. 10. Run program for specific primer on thermocycler. Gel Electrophoresis: 1. Add 1g of UltraPure™ Agarose to 100ml 1X TAE buffer. 2. Microwave for 1 minute or until melted. Caution that the mixture does not boil. Let the agarose cool, but not solidify. 3. Tape the open sides of the gel tray. Place combs into their slots. 4. Pour agarose into the tray and let solidify. 5. Peel the tape off the sides and remove combs. 6. Place the tray into a gel rig filled with 1X TAE buffer. 7. Aliquot 2l blue/yellow loading dye for every PCR sample onto a piece of Parafilm. 8. Pipet 8l PCR sample onto a dot of loading dye and mix by pipetting. 9. Pipet mixture into a well in the gel. 10. Repeat steps 8-9 until every PCR sample has been loaded into a well. 11. Replace cover on gel rig. Plug anode and cathode into the power source. Turn on (100 volts). 12. Set a timer for 50 minutes. 13. Turn off power source and unplug anode and cathode. Remove rig cover. 14. Remove gel from buffer and place in an ethidium bromide bath. CAUTION: Follow all important safety procedures when using ethidium bromide. 15. Set a timer for 20 minutes. 16. Remove gel from ethidium bromide bath. 17. Take a picture of the gel under UV light. Results Previous experimentation led to positive results of a positive horizontal Diapause - DNA from diapuse from a int13.2 female mated with an int12.1 male were extracted to determine if a Wolbachia transmission between int13.2 females transmission is seen in the offspring of an uninfected female and int12.1 males. Diapause DNA was horizontally by a male. screened to determine if this Sample # Type of Cross # of diapuase Result A35 4 transmission was being transferred to A12 4 the offspring of the host. Using a wsp A13 5 A22 6 primer, results suggest that Wolbachia A21 9 int12.1 Male x is not being inherited by the offspring A23 4 int13.2 Female A37 10 of the host, specifically in diapause. A46 4 A25 7 Positive and negative results are shown A27 3 in Figure 1. All experimental samples A18 4 B5 int13.2 xint13.2 6 from the cross between an int12.1 male C5 int12.1 x int12.1 1 + and an int13.2 female are negative for the presence of Wolbachia. The negative control crosses between an int13.2 male and an int13.2 female yields a negative result for the presence of Wolbachia. The positive control crosses between an int12.1 male and am int12.1 female yields a positive result. Figure 1 Figure 2 Embryo s from int13.2 Sample # Type of Cross # of embryos Result Sample # Type of Cross # of embryos Result females A6 7 A1 12 mated A8 9 A27 8 A9 4 A40 1 int12.1 Male x with A10 10 A23 int13.2 Female 7 Figure 1 A11 7 A44 5 int12.1 A14 6 A18 8 males A15 7 B1 5 int12.1 Male x int13.2 Male x A17 4 B3 10 were int13.2 Female int13.2 Female A19 11 B4 6 also A20 6 C4 2 A33 9 C6 3 + screene int12.1 Male x A42 10 C8 6 int12.1 Female d to A30 14 C9 6 A36 2 C5 2 determi A7 6 ne if the horizontal transmission seen in the cross was being inherited by the offspring of the host. DNA concentrations were determined because positive controls were not testing positive. Concentrations were really low, but similar to DNA from a positive embryo controls. 1l and 3l template DNA was used in PCR for the samples. An int12.1 DNA and an embryo DNA containing Wolbachia were used to compare the strength of bands in the tests. Experimental samples from the int13.2 female mated with the int12.1 male did not show any results suggesting the presence of Wolbachia. Negative control samples from the int13.2 female mated with the int13.2 male did not suggest a presence of Wolbachia either. Only 20% of the positive control samples suggested a presence of Wolbachia. The gel electrophoresis signal from the positive control was weak. If there was Wolbachia in any of the embryos, then the small signal of the sample was unlikely to show up. Embryos - DNA from embryos from an int13.2 female mated with an int12.1 male were extracted to determine if a Wolbachia transmission is seen in the offspring of an uninfected female transferred horizontally by an infected male. Figure 3 N. vitripennis - DNA from 13.2 females mated with 4.9 males to see if there is a transfer of Wolbachia from an infected male to an uninfected female Sample # Type of Cross Result A1 A2 A3 A4 A5 A6 A7 A8 + 4.9 Male x 13.2 A9 Female A10 A11 A12 A13 A14 A15 A16 A17 - Sample # Type of Cross Result A18 A19 A20 A21 4.9 Male x A22 13.2 Female A23 A24 A25 B1 + 4.9 Male x 4.9 B2 + Male B3 + C1 C2 13.2 Male x C3 13.2 Female C4 C5 - The positive results from the int13.2 females mated with the int12.1 males suggested the same patterns might be observed in other species of Nasonia with different strains of Wolbachia. Nasonia vitripennis 13.2 females were mated to 4,9 males and extracted to determine if the Wolbachia from the male was being transferred to the female. Experimental crosses between the 13.2 females and 4.9 males suggest that 4% of females become infected with Wolbachia (Figure 3). The mechanism of infection is unknown. Positive control crosses between 4.9 males and females suggest 100% infection rate. Negative control crosses between 13.2 males and females suggest 0% of samples were infected with Wolbachia. Figure 4 N. vitripennis - DNA from 13.2 females mated with 4.9 males was screened to determine if there is a horizontal transmission of Wolbachia from the male to the female. Each male was mated to 3 different females. Sample # Type of Cross Result 1.1 1.2 1.3 2.1 4.9 Male x 13.2 2.2 Female 2.3 3.1 3.2 3.3 1.4 + 4.9 Male x 4.9 1.5 + Female 1.6 7.1 13.2 Male x 7.2 13.2 Female 7.3 - Attempts to replicate Nasonia vitripennis data from Figure 3 proved unsuccessful. Males were mated to three different females to determine if Wolbachia would digress with each female. No Wolbachia was seen in 13.2 females mated with 4.9 males (Figure 4). Positive control crosses suggest a 66% transmission rate between 4.9 females mated with 4.9 males. Negative control crosses suggest a 0% transmission rate of Wolbachia between 13.2 females mated with 13.2 males. Another attempt to replicate data from Figure 3 will proceed in August/September 2008. Figure 5 N. giraulti - DNA from RV2R females mated with 16.2 males was screened to determine if there is a horizontal transmission of Wolbachia from the male to the female. Sample # Type of Cross Result A1 A2 A3 A4 A5 16.2 Male x A6 RV2R Female A7 A8 A9 A10 A11 B1 + 16.2 Male x B2 + 16.2 Female B3 + C1 RV2R Male x C2 RV2R Female C3 - Nasonia giraulti were also mated to determine if there was a transmission of Wolbachia from 16.2 males to RV2R females. Results, as shown in Figure 5, suggest that Wolbachia from a 16.2 male is not transferred to a RV2R female. Positive control crosses between a 16.2 male and female suggest 100% infection rate. Negative control crosses between a RV2R male and female suggest 0% infection rate. Figure 6 N. longicornis - DNA from females mated with males was screened to determine if there is a horizontal transmission of Wolbachia from the male to the female. Cross types cannot be determined at this date. Large amounts of unsuccessful courtships between Nasonia rose susupiscion about the male Nasonia strain. Virgin females were put on hosts to determine Nasonia strain and Wolbachia infection. Sample # A1 A2 A3 A4 A5 A6 A7 A8 A9 A10 A11 A12 A13 A14 A15 A16 A17 A18 A19 Result - Sample # A20 A21 A22 A23 A24 A25 A26 A27 A28 B1 B2 B3 B4 B5 C1 C2 C3 C4 C5 Result + - Nasonia longicornis females were mated to males. The type and infection status of the males is unknown. The positive strain was supposedly 2.1. The negative strain was supposedly IV7R3-1b. Many samples of the Nasonia longicornis were not mating with each other. Suspicion of the species of Nasonia was brought to attention. Wing size of the “2.1” males seemed too short to be Nasonia longicornis. In order to make a definitive conclusion, “2.1” virgin females were put on 2 hosts and honey. Wing size of these male offspring will be recorded once they emerge. Also, pupae from the hosts will be collected and stored in 95% ethanol until an extraction and amplification can be performed. These tests will conclude the strain of Wolbachia in the Nasonia. Results, as shown in Figure 6, suggest no Wolbachia was either transferred or transferred in the cross between the “2.1” male and the IV7R3-1b female. Results suggest that there may have been Wolbachia present in either the male or the female in the positive control crosses. All negative control crosses yielded results that suggest there is no Wolbachia present. Discussion Experimentation results suggest that Wolbachia is not inherited by the offspring, neither diapause nor embryos, of int13.2 females mated with int12.1 males, even though the Wolbachia is transferred from the infected male to the uninfected female. Also, horizontal transmission of Wolbachia has not been seen in N. giraulti or N. longicornis. A small infection rate was seen in N. vitripennis, but this could be due to human error. A definite conclusion will need to be drawn after more experimentation. Continued experimentation will consist of attempting to replicate results of the horizontal infection of Wolbachia in Nasonia vitripennis (13.2) females. Also, DNA from N. longicornis offspring will be analyzed to determine if they are infected with Wolbachia. These male offspring’s wing size will be analyzed to determine if they are in fact N. longicornis. Aside from the research of Wolbachia horizontal transmission in Nasonia, similar experiments will be conducted using Drosophila. References Clark, Michael E., Veneti, Z., Bourtzis, K., Karr, T.L. “The distribution and proliferation of the intracellular bacteria Wolbachia during spermatogenesis in Drosophila.” Mechanisms of Development. 111 (2002): 3-15. Riparbelli, M.G., Giordano, R., Callaini, G. “Effects of Wolbachia on sperm maturation and architecture in Drosophila simulans Riverside.” Mechanisms of Development. 124 (2007): 699-714. Tram, U., Fredrick, K., Werren, J.H., Sullivan, W. “Paternal chromosome segregation during the first mitotic division determines Wolbachia-induced cytoplasmic incompatibility phenotype.” Journal of Cell Science. 119 (2006): 3655-3663.