DOC - MRC Laboratory of Molecular Biology
advertisement
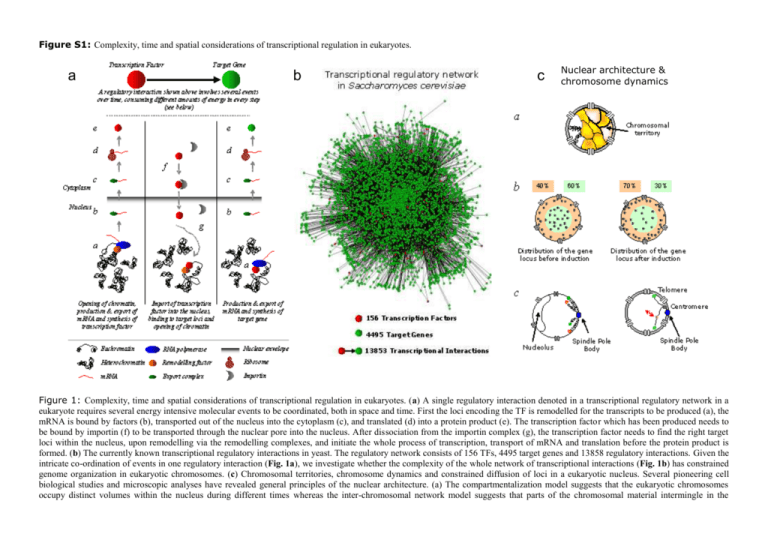
Figure S1: Complexity, time and spatial considerations of transcriptional regulation in eukaryotes. a b c Nuclear architecture & chromosome dynamics Figure 1: Complexity, time and spatial considerations of transcriptional regulation in eukaryotes. (a) A single regulatory interaction denoted in a transcriptional regulatory network in a eukaryote requires several energy intensive molecular events to be coordinated, both in space and time. First the loci encoding the TF is remodelled for the transcripts to be produced (a), the mRNA is bound by factors (b), transported out of the nucleus into the cytoplasm (c), and translated (d) into a protein product (e). The transcription factor which has been produced needs to be bound by importin (f) to be transported through the nuclear pore into the nucleus. After dissociation from the importin complex (g), the transcription factor needs to find the right target loci within the nucleus, upon remodelling via the remodelling complexes, and initiate the whole process of transcription, transport of mRNA and translation before the protein product is formed. (b) The currently known transcriptional regulatory interactions in yeast. The regulatory network consists of 156 TFs, 4495 target genes and 13858 regulatory interactions. Given the intricate co-ordination of events in one regulatory interaction (Fig. 1a), we investigate whether the complexity of the whole network of transcriptional interactions (Fig. 1b) has constrained genome organization in eukaryotic chromosomes. (c) Chromosomal territories, chromosome dynamics and constrained diffusion of loci in a eukaryotic nucleus. Several pioneering cell biological studies and microscopic analyses have revealed general principles of the nuclear architecture. (a) The compartmentalization model suggests that the eukaryotic chromosomes occupy distinct volumes within the nucleus during different times whereas the inter-chromosomal network model suggests that parts of the chromosomal material intermingle in the boundaries (1). In yeast, chromosomes can move in and out of the nearby territory and is represented as a gray area (adapted from (2)). (b) It has been recently shown that upon induction, the site of active transcription is brought closer to the nuclear envelope. The 2D projection of a nucleus is divided into two equal areas (light orange and light green). Before induction, the gene locus (gray dots) is roughly equally distributed (left) and upon activation, the loci preferentially moves towards the nuclear envelope (right). Adapted from (3). (c) Constrained movements and sites of anchoring chromosomes. Different regions of a chromosomal arm show a preference to reside closer or farther away from the center of the nucleus. The telomeres are normally tethered to the nuclear membrane, and the centromeres are closer to the spindle pole body, arranged in a Rabl-like conformation (left). Due to such tethering, loci with the chromosomal arm show constrained movement in the nuclear space (red arrow). Figure adapted from (2, 4). References: 1. 2. 3. 4. Branco MR & Pombo A (2006) Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol 4(5):e138. Gasser SM (2002) Visualizing chromatin dynamics in interphase nuclei. Science 296(5572):1412-1416. Cabal GG, et al. (2006) SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441(7094):770-773. Bystricky K, Laroche T, van Houwe G, Blaszczyk M, & Gasser SM (2005) Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J Cell Biol 168(3):375-387.