EMI_2282_sm_FigS1-TabS1-4
advertisement

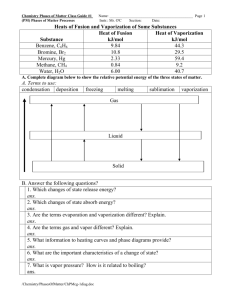
Table S1. In situ sulfate reduction rates (SRR) determined from a variety of production well and processing facility samples from two oil fields on Alaska’s NorthSlope. Table S2. Alkane-to-pristane or alkane-to-phytane peak area ratios for crude oil incubated in the presence (inoculated) or absence (sterile) of a methanogenic population enriched from a hot oilfield production well from Field B on Alaska’s North Slope. Bolded numbers show a decrease of the ratio in inoculated incubations relative to sterile incubations indicating that biodegradation has occurred. Alkane/Pristane Alkane/Phytane Alkane Sterile Inoculated Sterile Inoculated C10 0.874 0.490 1.034 0.558 C11 1.621 0.820 1.621 0.976 C12 1.647 1.083 1.949 1.287 C13 2.064 1.480 2.443 1.740 C14 1.884 1.501 2.229 1.835 C15 1.629 1.402 1.928 1.561 C16 1.511 1.366 1.789 1.547 C17 1.792 1.652 2.121 2.112 C18 1.226 1.180 1.451 1.38 C19 1.128 1.072 1.335 1.207 C20 1.058 1.057 1.252 1.197 able S3. Amounts (mol) of n-alkanes present in sterile and inoculated crude oilamended incubations enriched from production well fluids from Field B on Alaska’s North Slope and expected amounts (mol) of methane based on predicted stoichiometric reactions. n-alkane C# 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 mol nalkane, sterile control 0.229 0.316 0.452 0.453 0.496 0.634 0.570 0.510 0.963 0.521 0.657 0.715 0.689 0.711 0.716 0.700 0.836 0.778 0.728 0.631 0.623 0.614 0.508 mol nalkane, inoculated 1 0.102 0.161 0.242 0.256 0.295 0.379 0.338 0.283 0.547 0.224 0.216 0.203 0.188 0.185 0.217 0.272 0.348 0.374 0.391 0.395 0.394 0.386 0.321 total mol n-alkane consumed mol nalkane, inoculated 2 mol CH4 per mol n-alkane consumed mol CH4 theoretically expected 0.127 0.155 0.210 0.197 0.201 0.254 0.232 0.227 0.417 0.296 0.441 0.512 0.501 0.526 0.499 0.428 0.488 0.403 0.337 0.237 0.229 0.228 0.187 7 7.75 8.5 9.25 10 10.75 11.5 12.25 13 13.75 14.5 15.25 16 16.75 17.5 18.25 19 19.75 20.5 21.25 22 22.75 23.5 mol CH4 expected 0.886 1.201 1.783 1.824 2.010 2.735 2.662 2.783 5.416 4.074 6.397 7.815 8.017 8.811 8.740 7.818 9.263 7.969 6.912 5.027 5.048 5.198 4.399 7.33 116.79 Table S4. Summary of OTU in hydrocarbon-degrading methanogenic culture enriched from Field B. Primers1 Bacteria OTU % (Access. Total2 No.) BBACe11 29 (GU357467) Most similar sequences (Access. No.) Uncultured G55_D25_M_B_H02 (DQ997951) % identity (% query coverage)3 99 (98) Source Thermophilic anaerobic solid waste bioreactor ANS oil-water separator* Unpublished, not stated ANS oil processing facility coalescer* Mesothermic ANS oil well ANS oil well* Thermophilic anaerobic methanogenic reactor ANS oil well* Thermophilic microbial fuel cell Anaerobaculum hydrogeniformans OS1 99 (98) Thermotoga elfii strain G1 (DQ374393) 99 (100) CO1SHNF443 (FJ469358) 99 (100) 20 Uncultured D004011L07 (EU721778) 99 (99) Uncultured 2L1SGXO566 (FJ469295) 99 (87) 20 Thermacetogenium phaeum DSM 12270 98 (98) (NR_024688) 7 Uncultured 2L6SGXO407 (FJ469312) 99 (86) Uncultured Aminanaerobia clone SHBZ405 99 (99) (EU638937) Archaea BARCe03 100 Uncultured NAK-a1 (DQ867048) 99 (100) Natural gas field (GU357464) PS35SGXN478 (FJ44511) 99 (99) ANS oil-water separator* mcrA4 BMEg02 100 Uncultured PS130SGXI591 99 (100) ANS oil-water separator* (GU357469) Uncultured TUM-dMCRA-TR1-B1-5 92 (64) biogas reactor (FJ240435) Uncultured MDS-m-H03 (AB353242) 92 (58) mesophilic digested sludge Methanobacterium aarhusense (AY386125) 81 (98) marine sediment 5 mcrAA BMEg02AA 100 Uncultured PS1130SGXI591AA 100 (100) ANS oil-water separator* Uncultured TUM-dMCRA-TR1-B1-5 97 (64) biogas reactor (FJ240435) Uncultured MDS-m-H03 (AB353242) 96 (58) mesophilic digested sludge Methanobacterium aarhusense (AY386125) 91 (100) marine sediment 1 Primer sets used for each group are described in Experimental procedures 2 Bacteria, 41 sequences; Archaea, 46 sequences; mcrA, 23 sequences 3 % coverage of query sequence by database sequence, e.g. values < 100 indicate the database sequence being compared was shorter than the query sequence 4 Nucleotide sequence similarity 5 Amino acid similarity from deduced sequences * From the same ANS (Alaskan North Slope) oil field facility as the current enrichment BBACe04 (GU357465) BBACe02 (GU357463) BBACe03 (GU357464) BBACe10 (GU357466) 24 Fig. S1. Phylogenetic relationship of deduced methyl-coenzyme M reductase subunit A (mcrA) amino acid sequences from the Field B enrichment with respect to related sequences. The tree is constructed from approximately 219 amino acid positions (deduced from the mcrA nucleotide sequences) using the neighbor-joining algorithm. One thousand bootstrap replications were performed; only values greater than 700 are shown. The sequence shown in bold (BME09g02) and followed by an asterix is from the enrichment. It is representative of 23 sequences >99% similarity. Sequences in bold beginning with "PS" are from samples taken from the same field in 2006 (Duncan et al., 2009). Reference: Duncan, K. E., Gieg, L. M., Parisi, V. A., Tanner, R. S., Suflita, J. M., Green Tringe, S. and Bristow, J. (2009) Biocorrosive thermophilic microbial communities in Alaskan North Slope oil facilities. Environ Sci Technol 43: 7977-7984.